| p-value: | 1e-8 |
| log p-value: | -1.950e+01 |
| Information Content per bp: | 1.904 |
| Number of Target Sequences with motif | 268.0 |
| Percentage of Target Sequences with motif | 1.90% |
| Number of Background Sequences with motif | 150.4 |
| Percentage of Background Sequences with motif | 1.31% |
| Average Position of motif in Targets | 429.7 +/- 358.1bp |
| Average Position of motif in Background | 451.9 +/- 350.1bp |
| Strand Bias (log2 ratio + to - strand density) | 0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.02 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
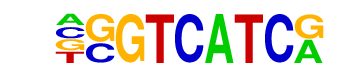
MA0089.1_NFE2L1::MafG/Jaspar
| Match Rank: | 1 |
| Score: | 0.86
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | GGTCATCR
-GTCATN- |
|


|
|
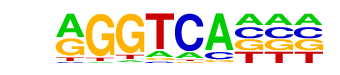
MF0004.1_Nuclear_Receptor_class/Jaspar
| Match Rank: | 2 |
| Score: | 0.72
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GGTCATCR
AGGTCA--- |
|
|
|
PB0108.1_Atf1_2/Jaspar
| Match Rank: | 3 |
| Score: | 0.70
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------GGTCATCR
NTTATTCGTCATNC |
|


|
|
MA0071.1_RORA_1/Jaspar
| Match Rank: | 4 |
| Score: | 0.68
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----GGTCATCR
ATCAAGGTCA--- |
|


|
|
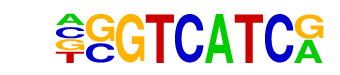
MF0002.1_bZIP_CREB/G-box-like_subclass/Jaspar
| Match Rank: | 5 |
| Score: | 0.68
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -GGTCATCR
ACGTCA--- |
|

|
|
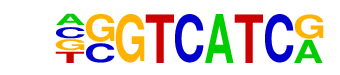
MA0117.1_Mafb/Jaspar
| Match Rank: | 6 |
| Score: | 0.68
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -GGTCATCR
NCGTCAGC- |
|

|
|
PB0038.1_Jundm2_1/Jaspar
| Match Rank: | 7 |
| Score: | 0.67
| | Offset: | -7
| | Orientation: | forward strand |
| Alignment: | -------GGTCATCR-
CCGATGACGTCATCGT |
|


|
|
Atf1(bZIP)/K562-ATF1-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 8 |
| Score: | 0.67
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---GGTCATCR
TGACGTCATC- |
|


|
|
Tbx20(T-box)/Heart-Tbx20-ChIP-Seq(GSE29636)/Homer
| Match Rank: | 9 |
| Score: | 0.66
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --GGTCATCR--
SCTGTCARCACC |
|


|
|
MafA(bZIP)/Islet-MafA-ChIP-Seq(GSE30298)/Homer
| Match Rank: | 10 |
| Score: | 0.66
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --GGTCATCR
TGAGTCAGCA |
|


|
|





















