| p-value: | 1e-7 |
| log p-value: | -1.742e+01 |
| Information Content per bp: | 1.955 |
| Number of Target Sequences with motif | 348.0 |
| Percentage of Target Sequences with motif | 2.47% |
| Number of Background Sequences with motif | 209.2 |
| Percentage of Background Sequences with motif | 1.82% |
| Average Position of motif in Targets | 433.1 +/- 347.6bp |
| Average Position of motif in Background | 434.9 +/- 354.6bp |
| Strand Bias (log2 ratio + to - strand density) | -0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.02 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
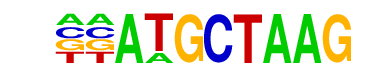
Oct4(POU/Homeobox)/mES-Oct4-ChIP-Seq(GSE11431)/Homer
| Match Rank: | 1 |
| Score: | 0.68
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --ATGCTAAG
TTATGCAAAT |
|

|
|
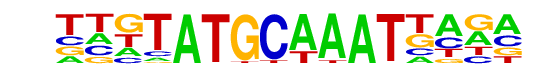
PH0145.1_Pou2f3/Jaspar
| Match Rank: | 2 |
| Score: | 0.67
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----ATGCTAAG----
TTGTATGCAAATTAGA |
|

|
|
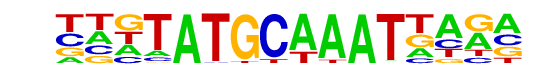
PH0144.1_Pou2f2/Jaspar
| Match Rank: | 3 |
| Score: | 0.67
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----ATGCTAAG----
TTGTATGCAAATTAGA |
|

|
|
Gata2(Zf)/K562-GATA2-ChIP-Seq(GSE18829)/Homer
| Match Rank: | 4 |
| Score: | 0.65
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | ATGCTAAG--
NAGATAAGNN |
|


|
|
Gata4(Zf)/Heart-Gata4-ChIP-Seq(GSE35151)/Homer
| Match Rank: | 5 |
| Score: | 0.64
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -ATGCTAAG-
NBWGATAAGR |
|


|
|
PB0054.1_Rfx3_1/Jaspar
| Match Rank: | 6 |
| Score: | 0.64
| | Offset: | -8
| | Orientation: | reverse strand |
| Alignment: | --------ATGCTAAG-------
NTNNNNNGTTGCTANGGNNCANA |
|


|
|
PB0055.1_Rfx4_1/Jaspar
| Match Rank: | 7 |
| Score: | 0.64
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----ATGCTAAG---
NNCGTTGCTATGGNN |
|


|
|
Gata1(Zf)/K562-GATA1-ChIP-Seq(GSE18829)/Homer
| Match Rank: | 8 |
| Score: | 0.64
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | ATGCTAAG--
CAGATAAGGN |
|


|
|
PB0022.1_Gata5_1/Jaspar
| Match Rank: | 9 |
| Score: | 0.64
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----ATGCTAAG-----
TAAACTGATAAGAAGAT |
|


|
|
PB0023.1_Gata6_1/Jaspar
| Match Rank: | 10 |
| Score: | 0.64
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----ATGCTAAG-----
TATAGAGATAAGAATTG |
|


|
|





















