| p-value: | 1e-12 |
| log p-value: | -2.882e+01 |
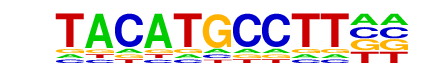
| Information Content per bp: | 1.530 |
| Number of Target Sequences with motif | 40.0 |
| Percentage of Target Sequences with motif | 0.17% |
| Number of Background Sequences with motif | 4.1 |
| Percentage of Background Sequences with motif | 0.04% |
| Average Position of motif in Targets | 446.0 +/- 258.5bp |
| Average Position of motif in Background | 727.9 +/- 322.7bp |
| Strand Bias (log2 ratio + to - strand density) | -0.6 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
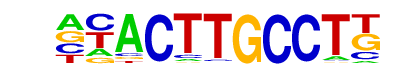
SD0002.1_at_AC_acceptor/Jaspar
| Match Rank: | 1 |
| Score: | 0.79
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TACATGCCTT
NNACTTGCCTT |
|

|
|
MA0090.1_TEAD1/Jaspar
| Match Rank: | 2 |
| Score: | 0.63
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | TACATGCCTT--
CACATTCCTCCG |
|

|
|
TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer
| Match Rank: | 3 |
| Score: | 0.62
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | TACATGCCTT-
-RCATTCCWGG |
|


|
|
HIF-1a(HLH)/MCF7-HIF1a-ChIP-Seq(GSE28352)/Homer
| Match Rank: | 4 |
| Score: | 0.62
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | TACATGCCTT
TACGTGCV-- |
|


|
|
PH0082.1_Irx2/Jaspar
| Match Rank: | 5 |
| Score: | 0.61
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----TACATGCCTT--
ANTNTTACATGTATNTA |
|


|
|
MA0032.1_FOXC1/Jaspar
| Match Rank: | 6 |
| Score: | 0.60
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TACATGCCTT
TACTNNNN-- |
|


|
|
PB0098.1_Zfp410_1/Jaspar
| Match Rank: | 7 |
| Score: | 0.60
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---TACATGCCTT----
NNNTCCATCCCATAANN |
|


|
|
TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer
| Match Rank: | 8 |
| Score: | 0.60
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | TACATGCCTT-
-GCATTCCAGN |
|


|
|
PH0084.1_Irx3_2/Jaspar
| Match Rank: | 9 |
| Score: | 0.60
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----TACATGCCTT---
AATATACATGTAATATA |
|


|
|
HIF2a(HLH)/785_O-HIF2a-ChIP-Seq(GSE34871)/Homer
| Match Rank: | 10 |
| Score: | 0.60
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---TACATGCCTT
GGGTACGTGC--- |
|


|
|





















