| p-value: | 1e-19 |
| log p-value: | -4.467e+01 |
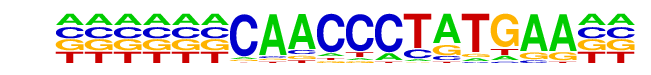
| Information Content per bp: | 1.615 |
| Number of Target Sequences with motif | 21.0 |
| Percentage of Target Sequences with motif | 6.40% |
| Number of Background Sequences with motif | 1.6 |
| Percentage of Background Sequences with motif | 0.56% |
| Average Position of motif in Targets | 436.9 +/- 374.8bp |
| Average Position of motif in Background | 806.8 +/- 490.1bp |
| Strand Bias (log2 ratio + to - strand density) | 2.2 |
| Multiplicity (# of sites on avg that occur together) | 1.10 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
MA0133.1_BRCA1/Jaspar
| Match Rank: | 1 |
| Score: | 0.64
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CAACCCTATGAA
ACAACAC------ |
|


|
|
PH0026.1_Duxbl/Jaspar
| Match Rank: | 2 |
| Score: | 0.60
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CAACCCTATGAA-----
CGACCCAATCAACGGTG |
|


|
|
CEBP:CEBP(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer
| Match Rank: | 3 |
| Score: | 0.57
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------CAACCCTATGAA--
NTNATGCAAYMNNHTGMAAY |
|

|
|
MA0074.1_RXRA::VDR/Jaspar
| Match Rank: | 4 |
| Score: | 0.56
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CAACCCTATGAA--
TGAACCCGATGACCC |
|


|
|
PH0057.1_Hoxb13/Jaspar
| Match Rank: | 5 |
| Score: | 0.55
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CAACCCTATGAA-----
-AACCCAATAAAATTCG |
|


|
|
TCFL2(HMG)/K562-TCF7L2-ChIP-Seq(GSE29196)/Homer
| Match Rank: | 6 |
| Score: | 0.55
| | Offset: | 4
| | Orientation: | reverse strand |
| Alignment: | CAACCCTATGAA--
----CCTTTGAWGT |
|


|
|
Tcf3(HMG)/mES-Tcf3-ChIP-Seq(GSE11724)/Homer
| Match Rank: | 7 |
| Score: | 0.54
| | Offset: | 4
| | Orientation: | reverse strand |
| Alignment: | CAACCCTATGAA--
----CCTTTGATGT |
|


|
|
Unknown(Homeobox)/Limb-p300-ChIP-Seq/Homer
| Match Rank: | 8 |
| Score: | 0.52
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | CAACCCTATGAA-
---NGCAATTAAA |
|


|
|
VDR(NR/DR3)/GM10855-VDR+vitD-ChIP-Seq(GSE22484)/Homer
| Match Rank: | 9 |
| Score: | 0.52
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----CAACCCTATGAA----
NNNTGAACTCNNTGACCTCN |
|


|
|
PB0128.1_Gcm1_2/Jaspar
| Match Rank: | 10 |
| Score: | 0.52
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---CAACCCTATGAA--
NTCNTCCCCTATNNGNN |
|


|
|





















