| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | p53(p53)/Saos-p53-ChIP-Seq(GSE15780)/Homer | 1e-3027 | -6.972e+03 | 0.0000 | 1173.0 | 30.19% | 0.3 | 0.01% | motif file (matrix) |
pdf |
| 2 |  | p53(p53)/Saos-p53-ChIP-Seq/Homer | 1e-3027 | -6.972e+03 | 0.0000 | 1173.0 | 30.19% | 0.3 | 0.01% | motif file (matrix) |
pdf |
| 3 |  | p63(p53)/Keratinocyte-p63-ChIP-Seq(GSE17611)/Homer | 1e-1686 | -3.884e+03 | 0.0000 | 1400.0 | 36.03% | 30.2 | 1.04% | motif file (matrix) |
pdf |
| 4 |  | p53(p53)/mES-cMyc-ChIP-Seq(GSE11431)/Homer | 1e-98 | -2.276e+02 | 0.0000 | 74.0 | 1.90% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 5 |  | Tcfcp2l1(CP2)/mES-Tcfcp2l1-ChIP-Seq(GSE11431)/Homer | 1e-31 | -7.324e+01 | 0.0000 | 69.0 | 1.78% | 8.7 | 0.30% | motif file (matrix) |
pdf |
| 6 |  | BMAL1(HLH)/Liver-Bmal1-ChIP-Seq(GSE39860)/Homer | 1e-29 | -6.864e+01 | 0.0000 | 525.0 | 13.51% | 233.0 | 8.07% | motif file (matrix) |
pdf |
| 7 |  | NRF1/Promoter/Homer | 1e-22 | -5.194e+01 | 0.0000 | 25.0 | 0.64% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 8 |  | GFX(?)/Promoter/Homer | 1e-20 | -4.612e+01 | 0.0000 | 23.0 | 0.59% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 9 |  | NPAS2(HLH)/Liver-NPAS2-ChIP-Seq(GSE39860)/Homer | 1e-18 | -4.299e+01 | 0.0000 | 228.0 | 5.87% | 89.1 | 3.09% | motif file (matrix) |
pdf |
| 10 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-17 | -4.120e+01 | 0.0000 | 98.0 | 2.52% | 26.7 | 0.92% | motif file (matrix) |
pdf |
| 11 |  | Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-16 | -3.914e+01 | 0.0000 | 46.0 | 1.18% | 8.0 | 0.28% | motif file (matrix) |
pdf |
| 12 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-15 | -3.580e+01 | 0.0000 | 95.0 | 2.44% | 28.0 | 0.97% | motif file (matrix) |
pdf |
| 13 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-14 | -3.417e+01 | 0.0000 | 82.0 | 2.11% | 22.1 | 0.77% | motif file (matrix) |
pdf |
| 14 |  | ETS(ETS)/Promoter/Homer | 1e-13 | -3.155e+01 | 0.0000 | 47.0 | 1.21% | 9.2 | 0.32% | motif file (matrix) |
pdf |
| 15 |  | GRHL2(CP2)/HBE-GRHL2-ChIP-Seq(GSE46194)/Homer | 1e-12 | -2.897e+01 | 0.0000 | 63.0 | 1.62% | 16.0 | 0.55% | motif file (matrix) |
pdf |
| 16 |  | AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-12 | -2.849e+01 | 0.0000 | 112.0 | 2.88% | 39.4 | 1.37% | motif file (matrix) |
pdf |
| 17 |  | AR-halfsite(NR)/LNCaP-AR-ChIP-Seq(GSE27824)/Homer | 1e-12 | -2.830e+01 | 0.0000 | 560.0 | 14.41% | 309.3 | 10.71% | motif file (matrix) |
pdf |
| 18 |  | Smad4(MAD)/ESC-SMAD4-ChIP-Seq(GSE29422)/Homer | 1e-10 | -2.368e+01 | 0.0000 | 250.0 | 6.43% | 121.7 | 4.21% | motif file (matrix) |
pdf |
| 19 |  | ZBTB33(Zf)/GM12878-ZBTB33-ChIP-Seq(GSE32465)/Homer | 1e-10 | -2.340e+01 | 0.0000 | 23.0 | 0.59% | 3.4 | 0.12% | motif file (matrix) |
pdf |
| 20 | 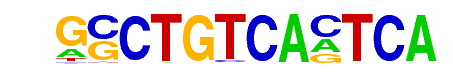 | PBX1(Homeobox)/MCF7-PBX1-ChIP-Seq(GSE28007)/Homer | 1e-9 | -2.230e+01 | 0.0000 | 14.0 | 0.36% | 1.3 | 0.05% | motif file (matrix) |
pdf |
| 21 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-9 | -2.089e+01 | 0.0000 | 66.0 | 1.70% | 21.1 | 0.73% | motif file (matrix) |
pdf |
| 22 |  | Smad3(MAD)/NPC-Smad3-ChIP-Seq(GSE36673)/Homer | 1e-9 | -2.083e+01 | 0.0000 | 376.0 | 9.68% | 204.5 | 7.08% | motif file (matrix) |
pdf |
| 23 |  | NF-E2(bZIP)/K562-NFE2-ChIP-Seq(GSE31477)/Homer | 1e-8 | -1.995e+01 | 0.0000 | 13.0 | 0.33% | 1.4 | 0.05% | motif file (matrix) |
pdf |
| 24 | 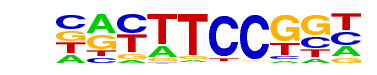 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-8 | -1.966e+01 | 0.0000 | 138.0 | 3.55% | 60.6 | 2.10% | motif file (matrix) |
pdf |
| 25 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-8 | -1.848e+01 | 0.0000 | 83.0 | 2.14% | 31.4 | 1.09% | motif file (matrix) |
pdf |
| 26 |  | Fox:Ebox(Forkhead:HLH)/Panc1-Foxa2-ChIP-Seq(GSE47459)/Homer | 1e-7 | -1.770e+01 | 0.0000 | 167.0 | 4.30% | 79.0 | 2.74% | motif file (matrix) |
pdf |
| 27 |  | NRF1(NRF)/MCF7-NRF1-ChIP-Seq(Unpublished)/Homer | 1e-7 | -1.767e+01 | 0.0000 | 12.0 | 0.31% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 28 |  | TRa(NR)/C17.2-TRa-ChIP-Seq(GSE38347)/Homer | 1e-7 | -1.759e+01 | 0.0000 | 58.0 | 1.49% | 19.4 | 0.67% | motif file (matrix) |
pdf |
| 29 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-7 | -1.688e+01 | 0.0000 | 75.0 | 1.93% | 28.7 | 1.00% | motif file (matrix) |
pdf |
| 30 |  | MafA(bZIP)/Islet-MafA-ChIP-Seq(GSE30298)/Homer | 1e-7 | -1.683e+01 | 0.0000 | 90.0 | 2.32% | 36.5 | 1.26% | motif file (matrix) |
pdf |
| 31 | 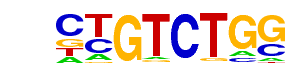 | Smad2(MAD)/ES-SMAD2-ChIP-Seq(GSE29422)/Homer | 1e-6 | -1.499e+01 | 0.0000 | 218.0 | 5.61% | 114.6 | 3.97% | motif file (matrix) |
pdf |
| 32 |  | JunD(bZIP)/K562-JunD-ChIP-Seq/Homer | 1e-6 | -1.384e+01 | 0.0000 | 14.0 | 0.36% | 2.6 | 0.09% | motif file (matrix) |
pdf |
| 33 |  | NFkB-p50,p52(RHD)/p50-ChIP-Chip(Schreiber et al.)/Homer | 1e-6 | -1.384e+01 | 0.0000 | 14.0 | 0.36% | 2.2 | 0.08% | motif file (matrix) |
pdf |
| 34 |  | Bach1(bZIP)/K562-Bach1-ChIP-Seq(GSE31477)/Homer | 1e-5 | -1.336e+01 | 0.0000 | 10.0 | 0.26% | 0.8 | 0.03% | motif file (matrix) |
pdf |
| 35 |  | Nrf2(bZIP)/Lymphoblast-Nrf2-ChIP-Seq(GSE37589)/Homer | 1e-4 | -1.134e+01 | 0.0001 | 9.0 | 0.23% | 1.1 | 0.04% | motif file (matrix) |
pdf |
| 36 | 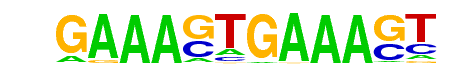 | IRF1(IRF)/PBMC-IRF1-ChIP-Seq(GSE43036)/Homer | 1e-4 | -1.058e+01 | 0.0002 | 12.0 | 0.31% | 2.9 | 0.10% | motif file (matrix) |
pdf |
| 37 |  | NFkB-p65(RHD)/GM12787-p65-ChIP-Seq(GSE19485)/Homer | 1e-4 | -9.604e+00 | 0.0004 | 36.0 | 0.93% | 13.7 | 0.47% | motif file (matrix) |
pdf |
| 38 |  | Six1(Homeobox)/Myoblast-Six1-ChIP-Chip(GSE20150)/Homer | 1e-4 | -9.583e+00 | 0.0004 | 34.0 | 0.87% | 13.0 | 0.45% | motif file (matrix) |
pdf |
| 39 |  | FOXA1:AR/LNCAP-AR-ChIP-Seq(GSE27824)/Homer | 1e-4 | -9.394e+00 | 0.0005 | 14.0 | 0.36% | 3.2 | 0.11% | motif file (matrix) |
pdf |
| 40 | 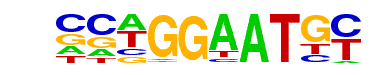 | TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer | 1e-3 | -9.166e+00 | 0.0006 | 85.0 | 2.19% | 41.3 | 1.43% | motif file (matrix) |
pdf |
| 41 |  | FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-3 | -8.438e+00 | 0.0013 | 180.0 | 4.63% | 102.9 | 3.56% | motif file (matrix) |
pdf |
| 42 |  | EBF1(EBF)/Near-E2A-ChIP-Seq(GSE21512)/Homer | 1e-3 | -8.367e+00 | 0.0014 | 98.0 | 2.52% | 50.0 | 1.73% | motif file (matrix) |
pdf |
| 43 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-3 | -8.270e+00 | 0.0015 | 101.0 | 2.60% | 52.8 | 1.83% | motif file (matrix) |
pdf |
| 44 |  | MyoD(HLH)/Myotube-MyoD-ChIP-Seq(GSE21614)/Homer | 1e-3 | -7.992e+00 | 0.0019 | 64.0 | 1.65% | 30.6 | 1.06% | motif file (matrix) |
pdf |
| 45 |  | GFY-Staf/Promoters/Homer | 1e-3 | -7.615e+00 | 0.0027 | 7.0 | 0.18% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 46 |  | OCT4-SOX2-TCF-NANOG((POU/Homeobox/HMG)/mES-Oct4-ChIP-Seq(GSE11431)/Homer | 1e-3 | -7.243e+00 | 0.0038 | 19.0 | 0.49% | 6.3 | 0.22% | motif file (matrix) |
pdf |
| 47 |  | Max(HLH)/K562-Max-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.198e+00 | 0.0039 | 125.0 | 3.22% | 69.7 | 2.41% | motif file (matrix) |
pdf |
| 48 |  | Bach2(bZIP)/OCILy7-Bach2-ChIP-Seq(GSE44420)/Homer | 1e-3 | -7.176e+00 | 0.0039 | 25.0 | 0.64% | 9.2 | 0.32% | motif file (matrix) |
pdf |
| 49 |  | CTCF(Zf)/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-3 | -6.924e+00 | 0.0049 | 12.0 | 0.31% | 3.8 | 0.13% | motif file (matrix) |
pdf |
| 50 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-2 | -6.866e+00 | 0.0051 | 176.0 | 4.53% | 103.1 | 3.57% | motif file (matrix) |
pdf |
| 51 |  | Arnt:Ahr(bHLH)/MCF7-Arnt-ChIP-Seq(Lo et al.)/Homer | 1e-2 | -6.815e+00 | 0.0053 | 94.0 | 2.42% | 50.8 | 1.76% | motif file (matrix) |
pdf |
| 52 |  | EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer | 1e-2 | -6.474e+00 | 0.0073 | 69.0 | 1.78% | 35.7 | 1.24% | motif file (matrix) |
pdf |
| 53 |  | HRE(HSF)/HepG2-HSF1-ChIP-Seq(GSE31477)/Homer | 1e-2 | -6.283e+00 | 0.0087 | 9.0 | 0.23% | 2.7 | 0.09% | motif file (matrix) |
pdf |
| 54 |  | FOXA1(Forkhead)/MCF7-FOXA1-ChIP-Seq(GSE26831)/Homer | 1e-2 | -6.252e+00 | 0.0088 | 148.0 | 3.81% | 86.3 | 2.99% | motif file (matrix) |
pdf |
| 55 |  | Foxa2(Forkhead)/Liver-Foxa2-ChIP-Seq(GSE25694)/Homer | 1e-2 | -6.084e+00 | 0.0102 | 123.0 | 3.17% | 70.5 | 2.44% | motif file (matrix) |
pdf |
| 56 |  | GFY(?)/Promoter/Homer | 1e-2 | -5.937e+00 | 0.0110 | 6.0 | 0.15% | 0.7 | 0.02% | motif file (matrix) |
pdf |
| 57 |  | GLI3(Zf)/GLI3-ChIP-Chip(GSE11077)/Homer | 1e-2 | -5.937e+00 | 0.0110 | 6.0 | 0.15% | 1.0 | 0.03% | motif file (matrix) |
pdf |
| 58 |  | Pax7-long(Paired/Homeobox)/Myoblast-Pax7-ChIP-Seq(GSE25064)/Homer | 1e-2 | -5.937e+00 | 0.0110 | 6.0 | 0.15% | 1.3 | 0.04% | motif file (matrix) |
pdf |
| 59 |  | Reverb(NR/DR2)/BLRP(RAW)-Reverba-ChIP-Seq(GSE45914)/Homer | 1e-2 | -5.937e+00 | 0.0110 | 6.0 | 0.15% | 1.7 | 0.06% | motif file (matrix) |
pdf |
| 60 |  | Tlx?(NR)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-2 | -5.801e+00 | 0.0124 | 34.0 | 0.87% | 15.5 | 0.54% | motif file (matrix) |
pdf |
| 61 |  | IRF2(IRF)/Erythroblas-IRF2-ChIP-Seq(GSE36985)/Homer | 1e-2 | -5.799e+00 | 0.0124 | 11.0 | 0.28% | 3.7 | 0.13% | motif file (matrix) |
pdf |
| 62 |  | Srebp2(HLH)/HepG2-Srebp2-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.799e+00 | 0.0124 | 11.0 | 0.28% | 3.5 | 0.12% | motif file (matrix) |
pdf |
| 63 |  | Rbpj1(?)/Panc1-Rbpj1-ChIP-Seq(GSE47459)/Homer | 1e-2 | -5.747e+00 | 0.0125 | 131.0 | 3.37% | 76.6 | 2.65% | motif file (matrix) |
pdf |
| 64 |  | Tcf12(HLH)/GM12878-Tcf12-ChIP-Seq(GSE32465)/Homer | 1e-2 | -5.702e+00 | 0.0128 | 83.0 | 2.14% | 45.3 | 1.57% | motif file (matrix) |
pdf |
| 65 |  | HRE(HSF)/Striatum-HSF1-ChIP-Seq(GSE38000)/Homer | 1e-2 | -5.595e+00 | 0.0141 | 13.0 | 0.33% | 4.3 | 0.15% | motif file (matrix) |
pdf |
| 66 |  | Pitx1(Homeobox)/Chicken-Pitx1-ChIP-Seq(GSE38910)/Homer | 1e-2 | -5.392e+00 | 0.0170 | 422.0 | 10.86% | 277.8 | 9.62% | motif file (matrix) |
pdf |
| 67 |  | Tbx20(T-box)/Heart-Tbx20-ChIP-Seq(GSE29636)/Homer | 1e-2 | -5.260e+00 | 0.0191 | 26.0 | 0.67% | 11.7 | 0.40% | motif file (matrix) |
pdf |
| 68 |  | RUNX1(Runt)/Jurkat-RUNX1-ChIP-Seq(GSE29180)/Homer | 1e-2 | -5.130e+00 | 0.0214 | 86.0 | 2.21% | 48.1 | 1.67% | motif file (matrix) |
pdf |
| 69 |  | ERE(NR/IR3)/MCF7-ERa-ChIP-Seq(Unpublished)/Homer | 1e-2 | -5.014e+00 | 0.0237 | 22.0 | 0.57% | 9.9 | 0.34% | motif file (matrix) |
pdf |
| 70 |  | ZFX(Zf)/mES-Zfx-ChIP-Seq(GSE11431)/Homer | 1e-2 | -4.815e+00 | 0.0285 | 117.0 | 3.01% | 69.8 | 2.42% | motif file (matrix) |
pdf |
| 71 |  | Rfx2(HTH)/LoVo-RFX2-ChIP-Seq(GSE49402)/Homer | 1e-2 | -4.755e+00 | 0.0298 | 10.0 | 0.26% | 3.2 | 0.11% | motif file (matrix) |
pdf |
| 72 |  | MafK(bZIP)/C2C12-MafK-ChIP-Seq(GSE36030)/Homer | 1e-2 | -4.725e+00 | 0.0303 | 16.0 | 0.41% | 6.6 | 0.23% | motif file (matrix) |
pdf |
| 73 |  | MyoG(HLH)/C2C12-MyoG-ChIP-Seq(GSE36024)/Homer | 1e-2 | -4.625e+00 | 0.0330 | 89.0 | 2.29% | 51.8 | 1.79% | motif file (matrix) |
pdf |

















































































































































