| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | p53(p53)/Saos-p53-ChIP-Seq(GSE15780)/Homer | 1e-1732 | -3.990e+03 | 0.0000 | 1206.0 | 25.59% | 13.5 | 0.42% | motif file (matrix) |
pdf |
| 2 |  | p53(p53)/Saos-p53-ChIP-Seq/Homer | 1e-1732 | -3.990e+03 | 0.0000 | 1206.0 | 25.59% | 13.5 | 0.42% | motif file (matrix) |
pdf |
| 3 |  | p63(p53)/Keratinocyte-p63-ChIP-Seq(GSE17611)/Homer | 1e-1204 | -2.774e+03 | 0.0000 | 1239.0 | 26.29% | 40.7 | 1.26% | motif file (matrix) |
pdf |
| 4 |  | p53(p53)/mES-cMyc-ChIP-Seq(GSE11431)/Homer | 1e-403 | -9.293e+02 | 0.0000 | 227.0 | 4.82% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 5 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-70 | -1.625e+02 | 0.0000 | 260.0 | 5.52% | 47.8 | 1.48% | motif file (matrix) |
pdf |
| 6 |  | GRHL2(CP2)/HBE-GRHL2-ChIP-Seq(GSE46194)/Homer | 1e-60 | -1.398e+02 | 0.0000 | 128.0 | 2.72% | 13.0 | 0.40% | motif file (matrix) |
pdf |
| 7 |  | AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-56 | -1.302e+02 | 0.0000 | 266.0 | 5.65% | 58.2 | 1.80% | motif file (matrix) |
pdf |
| 8 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-56 | -1.294e+02 | 0.0000 | 249.0 | 5.28% | 52.2 | 1.62% | motif file (matrix) |
pdf |
| 9 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-39 | -9.053e+01 | 0.0000 | 122.0 | 2.59% | 19.3 | 0.60% | motif file (matrix) |
pdf |
| 10 |  | Tcfcp2l1(CP2)/mES-Tcfcp2l1-ChIP-Seq(GSE11431)/Homer | 1e-37 | -8.647e+01 | 0.0000 | 87.0 | 1.85% | 11.0 | 0.34% | motif file (matrix) |
pdf |
| 11 |  | Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-35 | -8.256e+01 | 0.0000 | 123.0 | 2.61% | 21.1 | 0.65% | motif file (matrix) |
pdf |
| 12 | 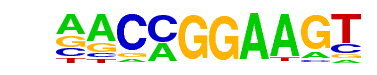 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-34 | -7.920e+01 | 0.0000 | 259.0 | 5.50% | 75.7 | 2.35% | motif file (matrix) |
pdf |
| 13 |  | ETS(ETS)/Promoter/Homer | 1e-31 | -7.212e+01 | 0.0000 | 75.0 | 1.59% | 9.1 | 0.28% | motif file (matrix) |
pdf |
| 14 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-30 | -7.026e+01 | 0.0000 | 137.0 | 2.91% | 30.0 | 0.93% | motif file (matrix) |
pdf |
| 15 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-29 | -6.855e+01 | 0.0000 | 172.0 | 3.65% | 43.5 | 1.35% | motif file (matrix) |
pdf |
| 16 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-28 | -6.504e+01 | 0.0000 | 151.0 | 3.20% | 36.5 | 1.13% | motif file (matrix) |
pdf |
| 17 |  | Smad4(MAD)/ESC-SMAD4-ChIP-Seq(GSE29422)/Homer | 1e-28 | -6.475e+01 | 0.0000 | 369.0 | 7.83% | 136.2 | 4.22% | motif file (matrix) |
pdf |
| 18 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-26 | -6.041e+01 | 0.0000 | 190.0 | 4.03% | 54.9 | 1.70% | motif file (matrix) |
pdf |
| 19 | 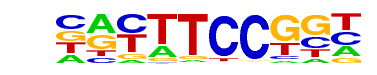 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-26 | -6.037e+01 | 0.0000 | 226.0 | 4.80% | 70.5 | 2.18% | motif file (matrix) |
pdf |
| 20 | 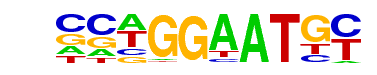 | TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer | 1e-23 | -5.475e+01 | 0.0000 | 206.0 | 4.37% | 64.6 | 2.00% | motif file (matrix) |
pdf |
| 21 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-21 | -4.873e+01 | 0.0000 | 267.0 | 5.67% | 97.3 | 3.01% | motif file (matrix) |
pdf |
| 22 |  | Bach2(bZIP)/OCILy7-Bach2-ChIP-Seq(GSE44420)/Homer | 1e-18 | -4.374e+01 | 0.0000 | 90.0 | 1.91% | 20.2 | 0.63% | motif file (matrix) |
pdf |
| 23 | 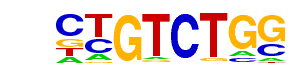 | Smad2(MAD)/ES-SMAD2-ChIP-Seq(GSE29422)/Homer | 1e-17 | -3.991e+01 | 0.0000 | 337.0 | 7.15% | 141.4 | 4.38% | motif file (matrix) |
pdf |
| 24 |  | TRa(NR)/C17.2-TRa-ChIP-Seq(GSE38347)/Homer | 1e-16 | -3.858e+01 | 0.0000 | 88.0 | 1.87% | 21.9 | 0.68% | motif file (matrix) |
pdf |
| 25 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-14 | -3.439e+01 | 0.0000 | 108.0 | 2.29% | 31.2 | 0.97% | motif file (matrix) |
pdf |
| 26 |  | RUNX(Runt)/HPC7-Runx1-ChIP-Seq(GSE22178)/Homer | 1e-14 | -3.226e+01 | 0.0000 | 101.0 | 2.14% | 29.4 | 0.91% | motif file (matrix) |
pdf |
| 27 |  | NRF1/Promoter/Homer | 1e-13 | -3.100e+01 | 0.0000 | 18.0 | 0.38% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 28 |  | PU.1(ETS)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-12 | -2.923e+01 | 0.0000 | 74.0 | 1.57% | 19.3 | 0.60% | motif file (matrix) |
pdf |
| 29 |  | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-12 | -2.777e+01 | 0.0000 | 194.0 | 4.12% | 77.6 | 2.40% | motif file (matrix) |
pdf |
| 30 |  | Rfx5(HTH)/GM12878-Rfx5-ChIP-Seq(GSE31477)/Homer | 1e-10 | -2.416e+01 | 0.0000 | 28.0 | 0.59% | 4.7 | 0.15% | motif file (matrix) |
pdf |
| 31 |  | MafA(bZIP)/Islet-MafA-ChIP-Seq(GSE30298)/Homer | 1e-9 | -2.154e+01 | 0.0000 | 115.0 | 2.44% | 42.8 | 1.33% | motif file (matrix) |
pdf |
| 32 |  | Smad3(MAD)/NPC-Smad3-ChIP-Seq(GSE36673)/Homer | 1e-9 | -2.139e+01 | 0.0000 | 535.0 | 11.35% | 282.6 | 8.75% | motif file (matrix) |
pdf |
| 33 |  | MafK(bZIP)/C2C12-MafK-ChIP-Seq(GSE36030)/Homer | 1e-8 | -1.954e+01 | 0.0000 | 52.0 | 1.10% | 14.5 | 0.45% | motif file (matrix) |
pdf |
| 34 |  | NF-E2(bZIP)/K562-NFE2-ChIP-Seq(GSE31477)/Homer | 1e-8 | -1.930e+01 | 0.0000 | 28.0 | 0.59% | 5.7 | 0.18% | motif file (matrix) |
pdf |
| 35 |  | NRF1(NRF)/MCF7-NRF1-ChIP-Seq(Unpublished)/Homer | 1e-8 | -1.901e+01 | 0.0000 | 13.0 | 0.28% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 36 |  | BMAL1(HLH)/Liver-Bmal1-ChIP-Seq(GSE39860)/Homer | 1e-8 | -1.853e+01 | 0.0000 | 378.0 | 8.02% | 193.4 | 5.99% | motif file (matrix) |
pdf |
| 37 |  | GATA3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-7 | -1.746e+01 | 0.0000 | 154.0 | 3.27% | 66.2 | 2.05% | motif file (matrix) |
pdf |
| 38 |  | SPDEF(ETS)/VCaP-SPDEF-ChIP-Seq(SRA014231)/Homer | 1e-7 | -1.739e+01 | 0.0000 | 152.0 | 3.23% | 65.5 | 2.03% | motif file (matrix) |
pdf |
| 39 |  | Gata4(Zf)/Heart-Gata4-ChIP-Seq(GSE35151)/Homer | 1e-7 | -1.717e+01 | 0.0000 | 108.0 | 2.29% | 43.0 | 1.33% | motif file (matrix) |
pdf |
| 40 |  | E2F7(E2F)/Hela-E2F7-ChIP-Seq(GSE32673)/Homer | 1e-7 | -1.681e+01 | 0.0000 | 12.0 | 0.25% | 1.1 | 0.03% | motif file (matrix) |
pdf |
| 41 |  | CEBP(bZIP)/CEBPb-ChIP-Seq(GSE21512)/Homer | 1e-6 | -1.583e+01 | 0.0000 | 94.0 | 1.99% | 36.3 | 1.12% | motif file (matrix) |
pdf |
| 42 |  | Nrf2(bZIP)/Lymphoblast-Nrf2-ChIP-Seq(GSE37589)/Homer | 1e-6 | -1.533e+01 | 0.0000 | 25.0 | 0.53% | 5.7 | 0.18% | motif file (matrix) |
pdf |
| 43 |  | RUNX1(Runt)/Jurkat-RUNX1-ChIP-Seq(GSE29180)/Homer | 1e-6 | -1.523e+01 | 0.0000 | 131.0 | 2.78% | 56.4 | 1.75% | motif file (matrix) |
pdf |
| 44 |  | Gata1(Zf)/K562-GATA1-ChIP-Seq(GSE18829)/Homer | 1e-6 | -1.471e+01 | 0.0000 | 64.0 | 1.36% | 22.8 | 0.71% | motif file (matrix) |
pdf |
| 45 |  | NPAS2(HLH)/Liver-NPAS2-ChIP-Seq(GSE39860)/Homer | 1e-6 | -1.402e+01 | 0.0000 | 177.0 | 3.76% | 83.7 | 2.59% | motif file (matrix) |
pdf |
| 46 |  | FOXA1(Forkhead)/MCF7-FOXA1-ChIP-Seq(GSE26831)/Homer | 1e-5 | -1.289e+01 | 0.0000 | 117.0 | 2.48% | 51.9 | 1.61% | motif file (matrix) |
pdf |
| 47 |  | FOXA1:AR/LNCAP-AR-ChIP-Seq(GSE27824)/Homer | 1e-5 | -1.265e+01 | 0.0000 | 10.0 | 0.21% | 0.5 | 0.02% | motif file (matrix) |
pdf |
| 48 |  | Gata2(Zf)/K562-GATA2-ChIP-Seq(GSE18829)/Homer | 1e-5 | -1.261e+01 | 0.0000 | 71.0 | 1.51% | 27.9 | 0.86% | motif file (matrix) |
pdf |
| 49 |  | Egr1(Zf)/K562-Egr1-ChIP-Seq(GSE32465)/Homer | 1e-5 | -1.205e+01 | 0.0000 | 74.0 | 1.57% | 29.6 | 0.92% | motif file (matrix) |
pdf |
| 50 |  | FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-5 | -1.187e+01 | 0.0000 | 147.0 | 3.12% | 69.6 | 2.15% | motif file (matrix) |
pdf |
| 51 |  | Bach1(bZIP)/K562-Bach1-ChIP-Seq(GSE31477)/Homer | 1e-5 | -1.170e+01 | 0.0000 | 22.0 | 0.47% | 5.7 | 0.18% | motif file (matrix) |
pdf |
| 52 |  | Egr2/Thymocytes-Egr2-ChIP-Seq(GSE34254)/Homer | 1e-4 | -1.133e+01 | 0.0001 | 13.0 | 0.28% | 2.5 | 0.08% | motif file (matrix) |
pdf |
| 53 |  | Fox:Ebox(Forkhead:HLH)/Panc1-Foxa2-ChIP-Seq(GSE47459)/Homer | 1e-4 | -1.087e+01 | 0.0001 | 118.0 | 2.50% | 54.8 | 1.70% | motif file (matrix) |
pdf |
| 54 |  | Maz(Zf)/HepG2-Maz-ChIP-Seq(GSE31477)/Homer | 1e-4 | -1.075e+01 | 0.0001 | 177.0 | 3.76% | 88.5 | 2.74% | motif file (matrix) |
pdf |
| 55 |  | HRE(HSF)/HepG2-HSF1-ChIP-Seq(GSE31477)/Homer | 1e-4 | -9.815e+00 | 0.0002 | 12.0 | 0.25% | 2.9 | 0.09% | motif file (matrix) |
pdf |
| 56 |  | X-box(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-4 | -9.815e+00 | 0.0002 | 12.0 | 0.25% | 2.6 | 0.08% | motif file (matrix) |
pdf |
| 57 |  | EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer | 1e-4 | -9.558e+00 | 0.0003 | 106.0 | 2.25% | 49.8 | 1.54% | motif file (matrix) |
pdf |
| 58 |  | PAX5(Paired/Homeobox)/GM12878-PAX5-ChIP-Seq(GSE32465)/Homer | 1e-4 | -9.497e+00 | 0.0003 | 58.0 | 1.23% | 23.3 | 0.72% | motif file (matrix) |
pdf |
| 59 |  | HIF-1a(HLH)/MCF7-HIF1a-ChIP-Seq(GSE28352)/Homer | 1e-3 | -9.032e+00 | 0.0005 | 31.0 | 0.66% | 10.4 | 0.32% | motif file (matrix) |
pdf |
| 60 |  | ELF5(ETS)/T47D-ELF5-ChIP-Seq(GSE30407)/Homer | 1e-3 | -9.003e+00 | 0.0005 | 94.0 | 1.99% | 43.0 | 1.33% | motif file (matrix) |
pdf |
| 61 |  | GATA-DR4(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-3 | -8.873e+00 | 0.0006 | 8.0 | 0.17% | 2.0 | 0.06% | motif file (matrix) |
pdf |
| 62 |  | NFkB-p50,p52(RHD)/p50-ChIP-Chip(Schreiber et al.)/Homer | 1e-3 | -8.873e+00 | 0.0006 | 8.0 | 0.17% | 0.4 | 0.01% | motif file (matrix) |
pdf |
| 63 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-3 | -8.730e+00 | 0.0006 | 41.0 | 0.87% | 15.4 | 0.48% | motif file (matrix) |
pdf |
| 64 |  | GFY-Staf/Promoters/Homer | 1e-3 | -8.376e+00 | 0.0009 | 11.0 | 0.23% | 3.0 | 0.09% | motif file (matrix) |
pdf |
| 65 |  | Pax8(Paired/Homeobox)/Thyroid-Pax8-ChIP-Seq(GSE26938)/Homer | 1e-3 | -7.647e+00 | 0.0018 | 49.0 | 1.04% | 20.6 | 0.64% | motif file (matrix) |
pdf |
| 66 | 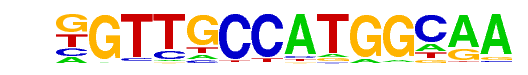 | Rfx1(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-3 | -7.576e+00 | 0.0019 | 27.0 | 0.57% | 9.7 | 0.30% | motif file (matrix) |
pdf |
| 67 |  | AP-2alpha(AP2)/Hela-AP2alpha-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.565e+00 | 0.0019 | 147.0 | 3.12% | 76.3 | 2.36% | motif file (matrix) |
pdf |
| 68 |  | AP-2gamma(AP2)/MCF7-TFAP2C-ChIP-Seq(GSE21234)/Homer | 1e-3 | -7.232e+00 | 0.0026 | 184.0 | 3.90% | 99.9 | 3.09% | motif file (matrix) |
pdf |
| 69 |  | AR-halfsite(NR)/LNCaP-AR-ChIP-Seq(GSE27824)/Homer | 1e-3 | -7.214e+00 | 0.0026 | 677.0 | 14.37% | 413.8 | 12.81% | motif file (matrix) |
pdf |
| 70 |  | PRDM14(Zf)/H1-PRDM14-ChIP-Seq(GSE22767)/Homer | 1e-3 | -7.194e+00 | 0.0026 | 20.0 | 0.42% | 6.6 | 0.20% | motif file (matrix) |
pdf |
| 71 |  | Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer | 1e-3 | -7.165e+00 | 0.0027 | 50.0 | 1.06% | 21.1 | 0.65% | motif file (matrix) |
pdf |
| 72 |  | IRF2(IRF)/Erythroblas-IRF2-ChIP-Seq(GSE36985)/Homer | 1e-3 | -7.021e+00 | 0.0031 | 10.0 | 0.21% | 2.5 | 0.08% | motif file (matrix) |
pdf |
| 73 |  | Chop(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer | 1e-2 | -6.761e+00 | 0.0039 | 30.0 | 0.64% | 11.7 | 0.36% | motif file (matrix) |
pdf |
| 74 |  | Foxa2(Forkhead)/Liver-Foxa2-ChIP-Seq(GSE25694)/Homer | 1e-2 | -6.757e+00 | 0.0039 | 85.0 | 1.80% | 41.8 | 1.29% | motif file (matrix) |
pdf |
| 75 |  | Arnt:Ahr(bHLH)/MCF7-Arnt-ChIP-Seq(Lo et al.)/Homer | 1e-2 | -6.580e+00 | 0.0046 | 88.0 | 1.87% | 43.1 | 1.33% | motif file (matrix) |
pdf |
| 76 |  | Klf4(Zf)/mES-Klf4-ChIP-Seq(GSE11431)/Homer | 1e-2 | -6.498e+00 | 0.0049 | 45.0 | 0.96% | 19.5 | 0.60% | motif file (matrix) |
pdf |
| 77 |  | Stat3(Stat)/mES-Stat3-ChIP-Seq(GSE11431)/Homer | 1e-2 | -6.066e+00 | 0.0074 | 64.0 | 1.36% | 30.6 | 0.95% | motif file (matrix) |
pdf |
| 78 |  | ETS:RUNX/Jurkat-RUNX1-ChIP-Seq(GSE17954)/Homer | 1e-2 | -5.867e+00 | 0.0088 | 14.0 | 0.30% | 4.6 | 0.14% | motif file (matrix) |
pdf |
| 79 |  | GLI3(Zf)/GLI3-ChIP-Chip(GSE11077)/Homer | 1e-2 | -5.867e+00 | 0.0088 | 14.0 | 0.30% | 4.6 | 0.14% | motif file (matrix) |
pdf |
| 80 |  | TATA-Box(TBP)/Promoter/Homer | 1e-2 | -5.655e+00 | 0.0108 | 109.0 | 2.31% | 57.8 | 1.79% | motif file (matrix) |
pdf |
| 81 |  | KLF5(Zf)/LoVo-KLF5-ChIP-Seq(GSE49402)/Homer | 1e-2 | -5.460e+00 | 0.0129 | 185.0 | 3.93% | 104.1 | 3.22% | motif file (matrix) |
pdf |
| 82 |  | Six1(Homeobox)/Myoblast-Six1-ChIP-Chip(GSE20150)/Homer | 1e-2 | -5.441e+00 | 0.0130 | 26.0 | 0.55% | 10.0 | 0.31% | motif file (matrix) |
pdf |
| 83 |  | RUNX2(Runt)/PCa-RUNX2-ChIP-Seq(GSE33889)/Homer | 1e-2 | -5.154e+00 | 0.0171 | 104.0 | 2.21% | 55.5 | 1.72% | motif file (matrix) |
pdf |
| 84 |  | c-Myc/mES-cMyc-ChIP-Seq(GSE11431)/Homer | 1e-2 | -5.083e+00 | 0.0182 | 65.0 | 1.38% | 32.2 | 1.00% | motif file (matrix) |
pdf |
| 85 |  | BORIS(Zf)/K562-CTCFL-ChIP-Seq(GSE32465)/Homer | 1e-2 | -4.937e+00 | 0.0208 | 29.0 | 0.62% | 12.5 | 0.39% | motif file (matrix) |
pdf |
| 86 |  | CEBP:CEBP(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer | 1e-2 | -4.818e+00 | 0.0231 | 15.0 | 0.32% | 5.2 | 0.16% | motif file (matrix) |
pdf |
| 87 |  | EBF1(EBF)/Near-E2A-ChIP-Seq(GSE21512)/Homer | 1e-2 | -4.767e+00 | 0.0241 | 145.0 | 3.08% | 81.9 | 2.54% | motif file (matrix) |
pdf |













































































































































































