| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-137 | -3.158e+02 | 0.0000 | 215.0 | 19.65% | 11.1 | 2.05% | motif file (matrix) |
pdf |
| 2 |  | AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-135 | -3.118e+02 | 0.0000 | 206.0 | 18.83% | 10.3 | 1.92% | motif file (matrix) |
pdf |
| 3 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-125 | -2.890e+02 | 0.0000 | 204.0 | 18.65% | 11.4 | 2.12% | motif file (matrix) |
pdf |
| 4 |  | RUNX1(Runt)/Jurkat-RUNX1-ChIP-Seq(GSE29180)/Homer | 1e-71 | -1.653e+02 | 0.0000 | 180.0 | 16.45% | 17.5 | 3.25% | motif file (matrix) |
pdf |
| 5 |  | Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-57 | -1.316e+02 | 0.0000 | 94.0 | 8.59% | 5.8 | 1.07% | motif file (matrix) |
pdf |
| 6 |  | Bach2(bZIP)/OCILy7-Bach2-ChIP-Seq(GSE44420)/Homer | 1e-56 | -1.292e+02 | 0.0000 | 54.0 | 4.94% | 1.0 | 0.18% | motif file (matrix) |
pdf |
| 7 |  | RUNX(Runt)/HPC7-Runx1-ChIP-Seq(GSE22178)/Homer | 1e-51 | -1.184e+02 | 0.0000 | 148.0 | 13.53% | 16.2 | 3.01% | motif file (matrix) |
pdf |
| 8 |  | STAT5(Stat)/mCD4+-Stat5-ChIP-Seq(GSE12346)/Homer | 1e-50 | -1.164e+02 | 0.0000 | 80.0 | 7.31% | 4.6 | 0.85% | motif file (matrix) |
pdf |
| 9 |  | RUNX2(Runt)/PCa-RUNX2-ChIP-Seq(GSE33889)/Homer | 1e-45 | -1.041e+02 | 0.0000 | 156.0 | 14.26% | 20.5 | 3.81% | motif file (matrix) |
pdf |
| 10 |  | STAT1(Stat)/HelaS3-STAT1-ChIP-Seq(GSE12782)/Homer | 1e-41 | -9.457e+01 | 0.0000 | 63.0 | 5.76% | 3.2 | 0.59% | motif file (matrix) |
pdf |
| 11 |  | RUNX-AML(Runt)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer | 1e-27 | -6.409e+01 | 0.0000 | 130.0 | 11.88% | 21.9 | 4.08% | motif file (matrix) |
pdf |
| 12 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-25 | -5.847e+01 | 0.0000 | 107.0 | 9.78% | 16.3 | 3.02% | motif file (matrix) |
pdf |
| 13 |  | Stat3+il21(Stat)/CD4-Stat3-ChIP-Seq(GSE19198)/Homer | 1e-24 | -5.730e+01 | 0.0000 | 82.0 | 7.50% | 10.0 | 1.86% | motif file (matrix) |
pdf |
| 14 | 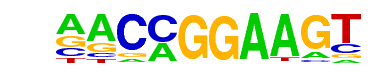 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-24 | -5.662e+01 | 0.0000 | 134.0 | 12.25% | 24.3 | 4.51% | motif file (matrix) |
pdf |
| 15 |  | STAT4(Stat)/CD4-Stat4-ChIP-Seq(GSE22104)/Homer | 1e-22 | -5.076e+01 | 0.0000 | 108.0 | 9.87% | 18.7 | 3.47% | motif file (matrix) |
pdf |
| 16 |  | Rfx2(HTH)/LoVo-RFX2-ChIP-Seq(GSE49402)/Homer | 1e-20 | -4.763e+01 | 0.0000 | 27.0 | 2.47% | 0.3 | 0.06% | motif file (matrix) |
pdf |
| 17 |  | NFkB-p65(RHD)/GM12787-p65-ChIP-Seq(GSE19485)/Homer | 1e-20 | -4.761e+01 | 0.0000 | 71.0 | 6.49% | 9.2 | 1.70% | motif file (matrix) |
pdf |
| 18 |  | EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer | 1e-20 | -4.760e+01 | 0.0000 | 87.0 | 7.95% | 13.7 | 2.54% | motif file (matrix) |
pdf |
| 19 |  | IRF4(IRF)/GM12878-IRF4-ChIP-Seq(GSE32465)/Homer | 1e-20 | -4.620e+01 | 0.0000 | 70.0 | 6.40% | 9.6 | 1.78% | motif file (matrix) |
pdf |
| 20 | 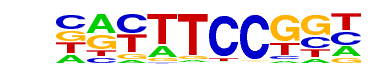 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-19 | -4.603e+01 | 0.0000 | 124.0 | 11.33% | 24.3 | 4.51% | motif file (matrix) |
pdf |
| 21 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-19 | -4.495e+01 | 0.0000 | 160.0 | 14.63% | 36.1 | 6.70% | motif file (matrix) |
pdf |
| 22 |  | bZIP:IRF/Th17-BatF-ChIP-Seq(GSE39756)/Homer | 1e-18 | -4.335e+01 | 0.0000 | 72.0 | 6.58% | 10.6 | 1.97% | motif file (matrix) |
pdf |
| 23 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-16 | -3.797e+01 | 0.0000 | 93.0 | 8.50% | 17.5 | 3.25% | motif file (matrix) |
pdf |
| 24 |  | PU.1(ETS)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-16 | -3.746e+01 | 0.0000 | 51.0 | 4.66% | 6.9 | 1.28% | motif file (matrix) |
pdf |
| 25 |  | RFX(HTH)/K562-RFX3-ChIP-Seq(SRA012198)/Homer | 1e-16 | -3.742e+01 | 0.0000 | 23.0 | 2.10% | 0.3 | 0.06% | motif file (matrix) |
pdf |
| 26 |  | Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer | 1e-15 | -3.678e+01 | 0.0000 | 46.0 | 4.20% | 5.3 | 0.98% | motif file (matrix) |
pdf |
| 27 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-15 | -3.567e+01 | 0.0000 | 66.0 | 6.03% | 10.1 | 1.88% | motif file (matrix) |
pdf |
| 28 |  | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-14 | -3.360e+01 | 0.0000 | 111.0 | 10.15% | 25.0 | 4.63% | motif file (matrix) |
pdf |
| 29 |  | NFAT(RHD)/Jurkat-NFATC1-ChIP-Seq(Jolma et al.)/Homer | 1e-13 | -3.047e+01 | 0.0000 | 92.0 | 8.41% | 19.4 | 3.61% | motif file (matrix) |
pdf |
| 30 |  | ELF5(ETS)/T47D-ELF5-ChIP-Seq(GSE30407)/Homer | 1e-12 | -2.970e+01 | 0.0000 | 61.0 | 5.58% | 10.8 | 2.00% | motif file (matrix) |
pdf |
| 31 |  | Stat3(Stat)/mES-Stat3-ChIP-Seq(GSE11431)/Homer | 1e-12 | -2.970e+01 | 0.0000 | 61.0 | 5.58% | 10.2 | 1.89% | motif file (matrix) |
pdf |
| 32 |  | ETS(ETS)/Promoter/Homer | 1e-12 | -2.894e+01 | 0.0000 | 45.0 | 4.11% | 6.5 | 1.21% | motif file (matrix) |
pdf |
| 33 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-11 | -2.686e+01 | 0.0000 | 62.0 | 5.67% | 11.4 | 2.12% | motif file (matrix) |
pdf |
| 34 |  | Hnf1(Homeobox)/Liver-Foxa2-Chip-Seq(GSE25694)/Homer | 1e-11 | -2.644e+01 | 0.0000 | 39.0 | 3.56% | 5.2 | 0.97% | motif file (matrix) |
pdf |
| 35 |  | NF-E2(bZIP)/K562-NFE2-ChIP-Seq(GSE31477)/Homer | 1e-11 | -2.565e+01 | 0.0000 | 18.0 | 1.65% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 36 |  | Usf2(HLH)/C2C12-Usf2-ChIP-Seq(GSE36030)/Homer | 1e-10 | -2.518e+01 | 0.0000 | 24.0 | 2.19% | 2.2 | 0.41% | motif file (matrix) |
pdf |
| 37 |  | SPDEF(ETS)/VCaP-SPDEF-ChIP-Seq(SRA014231)/Homer | 1e-10 | -2.506e+01 | 0.0000 | 92.0 | 8.41% | 21.6 | 4.01% | motif file (matrix) |
pdf |
| 38 | 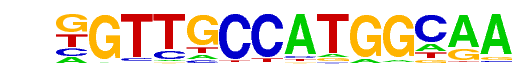 | Rfx1(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-10 | -2.492e+01 | 0.0000 | 29.0 | 2.65% | 3.4 | 0.64% | motif file (matrix) |
pdf |
| 39 |  | BMYB(HTH)/Hela-BMYB-ChIPSeq(GSE27030)/Homer | 1e-10 | -2.380e+01 | 0.0000 | 130.0 | 11.88% | 35.7 | 6.63% | motif file (matrix) |
pdf |
| 40 |  | Nrf2(bZIP)/Lymphoblast-Nrf2-ChIP-Seq(GSE37589)/Homer | 1e-10 | -2.345e+01 | 0.0000 | 17.0 | 1.55% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 41 |  | NFkB-p50,p52(RHD)/p50-ChIP-Chip(Schreiber et al.)/Homer | 1e-10 | -2.345e+01 | 0.0000 | 17.0 | 1.55% | 1.7 | 0.32% | motif file (matrix) |
pdf |
| 42 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-10 | -2.338e+01 | 0.0000 | 62.0 | 5.67% | 12.7 | 2.37% | motif file (matrix) |
pdf |
| 43 |  | Bach1(bZIP)/K562-Bach1-ChIP-Seq(GSE31477)/Homer | 1e-9 | -2.131e+01 | 0.0000 | 16.0 | 1.46% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 44 |  | MafK(bZIP)/C2C12-MafK-ChIP-Seq(GSE36030)/Homer | 1e-9 | -2.131e+01 | 0.0000 | 16.0 | 1.46% | 1.6 | 0.30% | motif file (matrix) |
pdf |
| 45 |  | Erra(NR)/HepG2-Erra-ChIP-Seq(GSE31477)/Homer | 1e-8 | -1.991e+01 | 0.0000 | 116.0 | 10.60% | 32.4 | 6.02% | motif file (matrix) |
pdf |
| 46 |  | NFAT:AP1/Jurkat-NFATC1-ChIP-Seq(Jolma et al.)/Homer | 1e-8 | -1.991e+01 | 0.0000 | 21.0 | 1.92% | 2.9 | 0.54% | motif file (matrix) |
pdf |
| 47 |  | X-box(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-8 | -1.991e+01 | 0.0000 | 21.0 | 1.92% | 2.6 | 0.48% | motif file (matrix) |
pdf |
| 48 |  | TRa(NR)/C17.2-TRa-ChIP-Seq(GSE38347)/Homer | 1e-8 | -1.879e+01 | 0.0000 | 25.0 | 2.29% | 3.8 | 0.70% | motif file (matrix) |
pdf |
| 49 |  | Foxa2(Forkhead)/Liver-Foxa2-ChIP-Seq(GSE25694)/Homer | 1e-7 | -1.764e+01 | 0.0000 | 65.0 | 5.94% | 15.4 | 2.85% | motif file (matrix) |
pdf |
| 50 |  | Pdx1(Homeobox)/Islet-Pdx1-ChIP-Seq(SRA008281)/Homer | 1e-7 | -1.764e+01 | 0.0000 | 65.0 | 5.94% | 15.2 | 2.82% | motif file (matrix) |
pdf |
| 51 |  | Hoxb4(Homeobox)/ES-Hoxb4-ChIP-Seq(GSE34014)/Homer | 1e-7 | -1.721e+01 | 0.0000 | 14.0 | 1.28% | 1.7 | 0.32% | motif file (matrix) |
pdf |
| 52 |  | NFkB-p65-Rel(RHD)/LPS-exp(GSE23622)/Homer | 1e-7 | -1.721e+01 | 0.0000 | 14.0 | 1.28% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 53 |  | ETS:RUNX/Jurkat-RUNX1-ChIP-Seq(GSE17954)/Homer | 1e-6 | -1.525e+01 | 0.0000 | 13.0 | 1.19% | 1.3 | 0.24% | motif file (matrix) |
pdf |
| 54 |  | Oct2(POU/Homeobox)/Bcell-Oct2-ChIP-Seq(GSE21512)/Homer | 1e-6 | -1.392e+01 | 0.0000 | 29.0 | 2.65% | 5.2 | 0.97% | motif file (matrix) |
pdf |
| 55 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-5 | -1.364e+01 | 0.0000 | 54.0 | 4.94% | 13.7 | 2.55% | motif file (matrix) |
pdf |
| 56 |  | Chop(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer | 1e-5 | -1.202e+01 | 0.0000 | 20.0 | 1.83% | 3.7 | 0.69% | motif file (matrix) |
pdf |
| 57 |  | CTCF(Zf)/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-5 | -1.158e+01 | 0.0000 | 11.0 | 1.01% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 58 |  | PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-4 | -1.090e+01 | 0.0001 | 110.0 | 10.05% | 36.1 | 6.71% | motif file (matrix) |
pdf |
| 59 |  | CEBP(bZIP)/CEBPb-ChIP-Seq(GSE21512)/Homer | 1e-4 | -9.908e+00 | 0.0002 | 46.0 | 4.20% | 12.9 | 2.40% | motif file (matrix) |
pdf |
| 60 |  | GLI3(Zf)/GLI3-ChIP-Chip(GSE11077)/Homer | 1e-4 | -9.866e+00 | 0.0002 | 10.0 | 0.91% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 61 |  | ISRE(IRF)/ThioMac-LPS-exp(GSE23622)/HOMER | 1e-4 | -9.866e+00 | 0.0002 | 10.0 | 0.91% | 1.5 | 0.27% | motif file (matrix) |
pdf |
| 62 |  | PRDM14(Zf)/H1-PRDM14-ChIP-Seq(GSE22767)/Homer | 1e-4 | -9.866e+00 | 0.0002 | 10.0 | 0.91% | 1.1 | 0.21% | motif file (matrix) |
pdf |
| 63 |  | BORIS(Zf)/K562-CTCFL-ChIP-Seq(GSE32465)/Homer | 1e-4 | -9.630e+00 | 0.0003 | 18.0 | 1.65% | 3.4 | 0.63% | motif file (matrix) |
pdf |
| 64 |  | Pitx1(Homeobox)/Chicken-Pitx1-ChIP-Seq(GSE38910)/Homer | 1e-3 | -8.701e+00 | 0.0006 | 260.0 | 23.77% | 104.4 | 19.39% | motif file (matrix) |
pdf |
| 65 |  | GATA3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-3 | -8.519e+00 | 0.0008 | 65.0 | 5.94% | 20.7 | 3.84% | motif file (matrix) |
pdf |
| 66 |  | NRF1(NRF)/MCF7-NRF1-ChIP-Seq(Unpublished)/Homer | 1e-3 | -8.245e+00 | 0.0010 | 9.0 | 0.82% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 67 |  | Smad4(MAD)/ESC-SMAD4-ChIP-Seq(GSE29422)/Homer | 1e-3 | -8.219e+00 | 0.0010 | 77.0 | 7.04% | 25.5 | 4.74% | motif file (matrix) |
pdf |
| 68 |  | Rfx5(HTH)/GM12878-Rfx5-ChIP-Seq(GSE31477)/Homer | 1e-3 | -8.138e+00 | 0.0011 | 35.0 | 3.20% | 9.1 | 1.68% | motif file (matrix) |
pdf |
| 69 |  | PRDM9(Zf)/Testis-DMC1-ChIP-Seq(GSE35498)/Homer | 1e-3 | -8.082e+00 | 0.0011 | 13.0 | 1.19% | 2.7 | 0.50% | motif file (matrix) |
pdf |
| 70 |  | RBPJ:Ebox/Panc1-Rbpj1-ChIP-Seq(GSE47459)/Homer | 1e-3 | -8.082e+00 | 0.0011 | 13.0 | 1.19% | 2.5 | 0.47% | motif file (matrix) |
pdf |
| 71 |  | NF1(CTF)/LNCAP-NF1-ChIP-Seq(Unpublished)/Homer | 1e-3 | -7.452e+00 | 0.0020 | 16.0 | 1.46% | 3.4 | 0.63% | motif file (matrix) |
pdf |
| 72 |  | CArG(MADS)/PUER-Srf-ChIP-Seq(Sullivan et al.)/Homer | 1e-3 | -7.452e+00 | 0.0020 | 16.0 | 1.46% | 3.2 | 0.60% | motif file (matrix) |
pdf |
| 73 |  | CEBP:AP1(bZIP)/ThioMac-CEBPb-ChIP-Seq(GSE21512)/Homer | 1e-3 | -7.272e+00 | 0.0023 | 60.0 | 5.48% | 19.5 | 3.62% | motif file (matrix) |
pdf |
| 74 |  | ZNF143|STAF(Zf)/CUTLL-ZNF143-ChIP-Seq(GSE29600)/Homer | 1e-3 | -7.199e+00 | 0.0025 | 19.0 | 1.74% | 4.8 | 0.89% | motif file (matrix) |
pdf |
| 75 |  | JunD(bZIP)/K562-JunD-ChIP-Seq/Homer | 1e-2 | -6.884e+00 | 0.0034 | 12.0 | 1.10% | 2.9 | 0.53% | motif file (matrix) |
pdf |
| 76 |  | TR4(NR/DR1)/Hela-TR4-ChIP-Seq(GSE24685)/Homer | 1e-2 | -6.726e+00 | 0.0039 | 8.0 | 0.73% | 1.7 | 0.32% | motif file (matrix) |
pdf |
| 77 |  | NFY(CCAAT)/Promoter/Homer | 1e-2 | -6.572e+00 | 0.0045 | 51.0 | 4.66% | 16.2 | 3.02% | motif file (matrix) |
pdf |
| 78 | 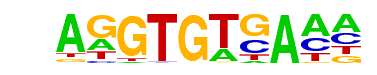 | Tbet(T-box)/CD8-Tbet-ChIP-Seq(GSE33802)/Homer | 1e-2 | -6.550e+00 | 0.0045 | 56.0 | 5.12% | 18.4 | 3.42% | motif file (matrix) |
pdf |
| 79 |  | Atf1(bZIP)/K562-ATF1-ChIP-Seq(GSE31477)/Homer | 1e-2 | -6.393e+00 | 0.0052 | 68.0 | 6.22% | 23.6 | 4.38% | motif file (matrix) |
pdf |
| 80 |  | Cdx2(Homeobox)/mES-Cdx2-ChIP-Seq(GSE14586)/Homer | 1e-2 | -6.153e+00 | 0.0065 | 32.0 | 2.93% | 9.4 | 1.74% | motif file (matrix) |
pdf |
| 81 |  | Mef2c(MADS)/GM12878-Mef2c-ChIP-Seq(GSE32465)/Homer | 1e-2 | -5.800e+00 | 0.0092 | 26.0 | 2.38% | 7.2 | 1.34% | motif file (matrix) |
pdf |
| 82 |  | HRE(HSF)/Striatum-HSF1-ChIP-Seq(GSE38000)/Homer | 1e-2 | -5.761e+00 | 0.0094 | 11.0 | 1.01% | 2.6 | 0.49% | motif file (matrix) |
pdf |
| 83 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-2 | -5.758e+00 | 0.0094 | 34.0 | 3.11% | 10.6 | 1.97% | motif file (matrix) |
pdf |
| 84 |  | STAT6/Macrophage-Stat6-ChIP-Seq(GSE38377)/Homer | 1e-2 | -5.626e+00 | 0.0106 | 54.0 | 4.94% | 18.1 | 3.37% | motif file (matrix) |
pdf |
| 85 |  | c-Jun-CRE(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.548e+00 | 0.0111 | 31.0 | 2.83% | 9.6 | 1.77% | motif file (matrix) |
pdf |
| 86 |  | Mef2a(MADS)/HL1-Mef2a.biotin-ChIP-Seq(GSE21529/Homer | 1e-2 | -5.548e+00 | 0.0111 | 31.0 | 2.83% | 9.4 | 1.74% | motif file (matrix) |
pdf |
| 87 |  | IRF2(IRF)/Erythroblas-IRF2-ChIP-Seq(GSE36985)/Homer | 1e-2 | -5.504e+00 | 0.0115 | 14.0 | 1.28% | 3.7 | 0.69% | motif file (matrix) |
pdf |
| 88 |  | PPARE(NR/DR1)/3T3L1-Pparg-ChIP-Seq(GSE13511)/Homer | 1e-2 | -5.369e+00 | 0.0130 | 41.0 | 3.75% | 13.7 | 2.55% | motif file (matrix) |
pdf |
| 89 |  | OCT4-SOX2-TCF-NANOG((POU/Homeobox/HMG)/mES-Oct4-ChIP-Seq(GSE11431)/Homer | 1e-2 | -5.318e+00 | 0.0136 | 7.0 | 0.64% | 1.9 | 0.35% | motif file (matrix) |
pdf |
| 90 |  | Bcl6(Zf)/Liver-Bcl6-ChIP-Seq(GSE31578)/Homer | 1e-2 | -5.275e+00 | 0.0140 | 98.0 | 8.96% | 37.3 | 6.93% | motif file (matrix) |
pdf |
| 91 |  | TATA-Box(TBP)/Promoter/Homer | 1e-2 | -5.252e+00 | 0.0142 | 58.0 | 5.30% | 20.5 | 3.80% | motif file (matrix) |
pdf |
| 92 |  | FOXA1(Forkhead)/MCF7-FOXA1-ChIP-Seq(GSE26831)/Homer | 1e-2 | -5.174e+00 | 0.0151 | 65.0 | 5.94% | 23.2 | 4.32% | motif file (matrix) |
pdf |
| 93 |  | Sox6(HMG)/Myotubes-Sox6-ChIP-Seq(GSE32627)/Homer | 1e-2 | -5.148e+00 | 0.0154 | 93.0 | 8.50% | 35.9 | 6.67% | motif file (matrix) |
pdf |
| 94 |  | MYB(HTH)/ERMYB-Myb-ChIPSeq(GSE22095)/Homer | 1e-2 | -4.864e+00 | 0.0202 | 135.0 | 12.34% | 54.0 | 10.04% | motif file (matrix) |
pdf |



























































































































































































