| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | Bach1(bZIP)/K562-Bach1-ChIP-Seq(GSE31477)/Homer | 1e-6 | -1.564e+01 | 0.0000 | 87.0 | 0.69% | 62.6 | 0.38% | motif file (matrix) |
pdf |
| 2 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-6 | -1.508e+01 | 0.0000 | 823.0 | 6.50% | 903.5 | 5.46% | motif file (matrix) |
pdf |
| 3 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-6 | -1.416e+01 | 0.0001 | 2161.0 | 17.06% | 2563.6 | 15.49% | motif file (matrix) |
pdf |
| 4 |  | PU.1(ETS)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-5 | -1.297e+01 | 0.0001 | 612.0 | 4.83% | 663.8 | 4.01% | motif file (matrix) |
pdf |
| 5 | 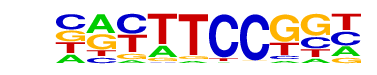 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-5 | -1.277e+01 | 0.0001 | 1318.0 | 10.40% | 1525.8 | 9.22% | motif file (matrix) |
pdf |
| 6 |  | Sp1(Zf)/Promoter/Homer | 1e-5 | -1.276e+01 | 0.0001 | 134.0 | 1.06% | 115.1 | 0.70% | motif file (matrix) |
pdf |
| 7 |  | Nrf2(bZIP)/Lymphoblast-Nrf2-ChIP-Seq(GSE37589)/Homer | 1e-5 | -1.269e+01 | 0.0001 | 78.0 | 0.62% | 58.2 | 0.35% | motif file (matrix) |
pdf |
| 8 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-4 | -1.086e+01 | 0.0006 | 800.0 | 6.31% | 904.6 | 5.46% | motif file (matrix) |
pdf |
| 9 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-4 | -1.049e+01 | 0.0008 | 714.0 | 5.64% | 802.1 | 4.85% | motif file (matrix) |
pdf |
| 10 |  | MafF(bZIP)/HepG2-MafF-ChIP-Seq(GSE31477)/Homer | 1e-4 | -1.021e+01 | 0.0009 | 363.0 | 2.87% | 383.6 | 2.32% | motif file (matrix) |
pdf |
| 11 |  | CTCF(Zf)/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-4 | -9.612e+00 | 0.0015 | 141.0 | 1.11% | 131.1 | 0.79% | motif file (matrix) |
pdf |
| 12 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-3 | -8.785e+00 | 0.0031 | 1298.0 | 10.25% | 1539.2 | 9.30% | motif file (matrix) |
pdf |
| 13 | 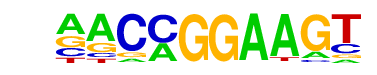 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-3 | -8.542e+00 | 0.0037 | 1704.0 | 13.45% | 2052.3 | 12.40% | motif file (matrix) |
pdf |
| 14 |  | HIF-1a(HLH)/MCF7-HIF1a-ChIP-Seq(GSE28352)/Homer | 1e-3 | -8.126e+00 | 0.0052 | 214.0 | 1.69% | 219.5 | 1.33% | motif file (matrix) |
pdf |
| 15 |  | Tcf12(HLH)/GM12878-Tcf12-ChIP-Seq(GSE32465)/Homer | 1e-3 | -8.004e+00 | 0.0055 | 1180.0 | 9.31% | 1400.0 | 8.46% | motif file (matrix) |
pdf |
| 16 |  | NF-E2(bZIP)/K562-NFE2-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.937e+00 | 0.0055 | 87.0 | 0.69% | 77.6 | 0.47% | motif file (matrix) |
pdf |
| 17 |  | Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.866e+00 | 0.0056 | 278.0 | 2.19% | 295.6 | 1.79% | motif file (matrix) |
pdf |
| 18 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-3 | -7.458e+00 | 0.0079 | 1084.0 | 8.56% | 1286.1 | 7.77% | motif file (matrix) |
pdf |
| 19 |  | EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer | 1e-3 | -7.198e+00 | 0.0097 | 964.0 | 7.61% | 1139.8 | 6.89% | motif file (matrix) |
pdf |
| 20 |  | NFkB-p65-Rel(RHD)/LPS-exp(GSE23622)/Homer | 1e-3 | -7.184e+00 | 0.0097 | 53.0 | 0.42% | 43.8 | 0.26% | motif file (matrix) |
pdf |
| 21 |  | CEBP:CEBP(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer | 1e-3 | -7.080e+00 | 0.0099 | 172.0 | 1.36% | 175.9 | 1.06% | motif file (matrix) |
pdf |
| 22 |  | MafK(bZIP)/C2C12-MafK-ChIP-Seq(GSE36030)/Homer | 1e-2 | -6.797e+00 | 0.0125 | 279.0 | 2.20% | 302.0 | 1.82% | motif file (matrix) |
pdf |
| 23 |  | AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-2 | -6.267e+00 | 0.0203 | 879.0 | 6.94% | 1043.1 | 6.30% | motif file (matrix) |
pdf |
| 24 |  | E2A(HLH)/proBcell-E2A-ChIP-Seq(GSE21978)/Homer | 1e-2 | -5.985e+00 | 0.0258 | 1646.0 | 12.99% | 2014.2 | 12.17% | motif file (matrix) |
pdf |
| 25 | 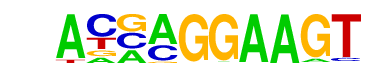 | ELF5(ETS)/T47D-ELF5-ChIP-Seq(GSE30407)/Homer | 1e-2 | -5.670e+00 | 0.0339 | 931.0 | 7.35% | 1115.8 | 6.74% | motif file (matrix) |
pdf |
| 26 |  | BORIS(Zf)/K562-CTCFL-ChIP-Seq(GSE32465)/Homer | 1e-2 | -5.527e+00 | 0.0376 | 177.0 | 1.40% | 188.1 | 1.14% | motif file (matrix) |
pdf |
| 27 |  | ETS:RUNX/Jurkat-RUNX1-ChIP-Seq(GSE17954)/Homer | 1e-2 | -5.131e+00 | 0.0539 | 84.0 | 0.66% | 82.8 | 0.50% | motif file (matrix) |
pdf |
| 28 |  | Ap4(HLH)/AML-Tfap4-ChIP-Seq(GSE45738)/Homer | 1e-2 | -5.029e+00 | 0.0575 | 1509.0 | 11.91% | 1855.8 | 11.21% | motif file (matrix) |
pdf |
| 29 |  | Gata1(Zf)/K562-GATA1-ChIP-Seq(GSE18829)/Homer | 1e-2 | -5.002e+00 | 0.0575 | 722.0 | 5.70% | 861.1 | 5.20% | motif file (matrix) |
pdf |
| 30 |  | Myf5(bHLH)/GM-Myf5-ChIP-Seq(GSE24852)/Homer | 1e-2 | -4.954e+00 | 0.0579 | 869.0 | 6.86% | 1046.1 | 6.32% | motif file (matrix) |
pdf |
| 31 |  | Bach2(bZIP)/OCILy7-Bach2-ChIP-Seq(GSE44420)/Homer | 1e-2 | -4.857e+00 | 0.0617 | 220.0 | 1.74% | 243.8 | 1.47% | motif file (matrix) |
pdf |
| 32 |  | AP-2alpha(AP2)/Hela-AP2alpha-ChIP-Seq(GSE31477)/Homer | 1e-2 | -4.746e+00 | 0.0668 | 726.0 | 5.73% | 869.9 | 5.25% | motif file (matrix) |
pdf |
| 33 |  | EBF1(EBF)/Near-E2A-ChIP-Seq(GSE21512)/Homer | 1e-2 | -4.743e+00 | 0.0668 | 1140.0 | 9.00% | 1391.1 | 8.40% | motif file (matrix) |
pdf |

































































