| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | Olig2(bHLH)/Neuron-Olig2-ChIP-Seq(GSE30882)/Homer | 1e-11 | -2.760e+01 | 0.0000 | 4504.0 | 20.66% | 2931.1 | 18.78% | motif file (matrix) |
pdf |
| 2 |  | Atoh1(bHLH)/Cerebellum-Atoh1-ChIP-Seq(GSE22111)/Homer | 1e-8 | -1.975e+01 | 0.0000 | 2339.0 | 10.73% | 1490.6 | 9.55% | motif file (matrix) |
pdf |
| 3 |  | MyoD(HLH)/Myotube-MyoD-ChIP-Seq(GSE21614)/Homer | 1e-8 | -1.924e+01 | 0.0000 | 1448.0 | 6.64% | 892.7 | 5.72% | motif file (matrix) |
pdf |
| 4 | 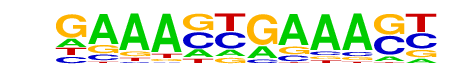 | IRF2(IRF)/Erythroblas-IRF2-ChIP-Seq(GSE36985)/Homer | 1e-8 | -1.861e+01 | 0.0000 | 227.0 | 1.04% | 109.4 | 0.70% | motif file (matrix) |
pdf |
| 5 |  | ISRE(IRF)/ThioMac-LPS-exp(GSE23622)/HOMER | 1e-7 | -1.800e+01 | 0.0000 | 145.0 | 0.67% | 63.8 | 0.41% | motif file (matrix) |
pdf |
| 6 |  | AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-7 | -1.648e+01 | 0.0000 | 1563.0 | 7.17% | 981.4 | 6.29% | motif file (matrix) |
pdf |
| 7 |  | KLF5(Zf)/LoVo-KLF5-ChIP-Seq(GSE49402)/Homer | 1e-6 | -1.547e+01 | 0.0000 | 1704.0 | 7.82% | 1081.4 | 6.93% | motif file (matrix) |
pdf |
| 8 |  | SCL(HLH)/HPC7-Scl-ChIP-Seq(GSE13511)/Homer | 1e-6 | -1.526e+01 | 0.0000 | 9747.0 | 44.72% | 6715.6 | 43.03% | motif file (matrix) |
pdf |
| 9 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-6 | -1.514e+01 | 0.0000 | 1368.0 | 6.28% | 856.8 | 5.49% | motif file (matrix) |
pdf |
| 10 |  | Tcf12(HLH)/GM12878-Tcf12-ChIP-Seq(GSE32465)/Homer | 1e-6 | -1.464e+01 | 0.0000 | 1795.0 | 8.24% | 1147.7 | 7.35% | motif file (matrix) |
pdf |
| 11 |  | Myf5(bHLH)/GM-Myf5-ChIP-Seq(GSE24852)/Homer | 1e-6 | -1.453e+01 | 0.0000 | 1346.0 | 6.18% | 844.0 | 5.41% | motif file (matrix) |
pdf |
| 12 |  | Ap4(HLH)/AML-Tfap4-ChIP-Seq(GSE45738)/Homer | 1e-5 | -1.336e+01 | 0.0000 | 2394.0 | 10.98% | 1564.1 | 10.02% | motif file (matrix) |
pdf |
| 13 |  | EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer | 1e-5 | -1.228e+01 | 0.0001 | 1632.0 | 7.49% | 1049.5 | 6.72% | motif file (matrix) |
pdf |
| 14 |  | Ptf1a(HLH)/Panc1-Ptf1a-ChIP-Seq(GSE47459)/Homer | 1e-5 | -1.202e+01 | 0.0001 | 5279.0 | 24.22% | 3584.2 | 22.96% | motif file (matrix) |
pdf |
| 15 |  | PU.1(ETS)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-5 | -1.195e+01 | 0.0001 | 953.0 | 4.37% | 592.8 | 3.80% | motif file (matrix) |
pdf |
| 16 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-4 | -1.047e+01 | 0.0004 | 3392.0 | 15.56% | 2277.0 | 14.59% | motif file (matrix) |
pdf |
| 17 |  | EKLF(Zf)/Erythrocyte-Klf1-ChIP-Seq(GSE20478)/Homer | 1e-4 | -1.043e+01 | 0.0004 | 327.0 | 1.50% | 186.7 | 1.20% | motif file (matrix) |
pdf |
| 18 |  | NeuroD1(bHLH)/Islet-NeuroD1-ChIP-Seq(GSE30298)/Homer | 1e-4 | -1.013e+01 | 0.0005 | 1733.0 | 7.95% | 1131.5 | 7.25% | motif file (matrix) |
pdf |
| 19 |  | Pitx1(Homeobox)/Chicken-Pitx1-ChIP-Seq(GSE38910)/Homer | 1e-4 | -1.011e+01 | 0.0005 | 9511.0 | 43.63% | 6604.5 | 42.31% | motif file (matrix) |
pdf |
| 20 |  | Bach2(bZIP)/OCILy7-Bach2-ChIP-Seq(GSE44420)/Homer | 1e-4 | -9.852e+00 | 0.0006 | 379.0 | 1.74% | 221.9 | 1.42% | motif file (matrix) |
pdf |
| 21 |  | BORIS(Zf)/K562-CTCFL-ChIP-Seq(GSE32465)/Homer | 1e-3 | -8.567e+00 | 0.0022 | 230.0 | 1.06% | 130.0 | 0.83% | motif file (matrix) |
pdf |
| 22 |  | Sp1(Zf)/Promoter/Homer | 1e-3 | -8.476e+00 | 0.0023 | 161.0 | 0.74% | 86.2 | 0.55% | motif file (matrix) |
pdf |
| 23 |  | MyoG(HLH)/C2C12-MyoG-ChIP-Seq(GSE36024)/Homer | 1e-3 | -8.317e+00 | 0.0026 | 1967.0 | 9.02% | 1305.4 | 8.36% | motif file (matrix) |
pdf |
| 24 |  | CTCF(Zf)/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-3 | -8.277e+00 | 0.0026 | 202.0 | 0.93% | 112.1 | 0.72% | motif file (matrix) |
pdf |
| 25 |  | NFY(CCAAT)/Promoter/Homer | 1e-3 | -8.111e+00 | 0.0030 | 1361.0 | 6.24% | 889.6 | 5.70% | motif file (matrix) |
pdf |
| 26 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-3 | -7.988e+00 | 0.0032 | 1126.0 | 5.17% | 729.2 | 4.67% | motif file (matrix) |
pdf |
| 27 |  | Nrf2(bZIP)/Lymphoblast-Nrf2-ChIP-Seq(GSE37589)/Homer | 1e-3 | -7.869e+00 | 0.0035 | 95.0 | 0.44% | 47.5 | 0.30% | motif file (matrix) |
pdf |
| 28 |  | c-Jun-CRE(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.328e+00 | 0.0058 | 555.0 | 2.55% | 346.2 | 2.22% | motif file (matrix) |
pdf |
| 29 |  | Klf4(Zf)/mES-Klf4-ChIP-Seq(GSE11431)/Homer | 1e-3 | -7.236e+00 | 0.0061 | 493.0 | 2.26% | 305.2 | 1.96% | motif file (matrix) |
pdf |
| 30 |  | bHLHE40(HLH)/HepG2-BHLHE40-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.216e+00 | 0.0061 | 407.0 | 1.87% | 248.6 | 1.59% | motif file (matrix) |
pdf |
| 31 | 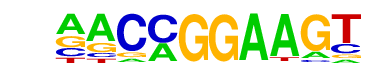 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-3 | -7.182e+00 | 0.0061 | 2695.0 | 12.36% | 1821.6 | 11.67% | motif file (matrix) |
pdf |
| 32 |  | NF-E2(bZIP)/K562-NFE2-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.104e+00 | 0.0063 | 121.0 | 0.56% | 64.6 | 0.41% | motif file (matrix) |
pdf |
| 33 | 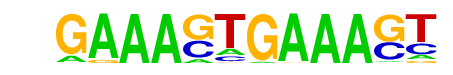 | IRF1(IRF)/PBMC-IRF1-ChIP-Seq(GSE43036)/Homer | 1e-3 | -7.097e+00 | 0.0063 | 286.0 | 1.31% | 169.6 | 1.09% | motif file (matrix) |
pdf |
| 34 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-2 | -6.834e+00 | 0.0078 | 2033.0 | 9.33% | 1363.6 | 8.74% | motif file (matrix) |
pdf |
| 35 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-2 | -6.821e+00 | 0.0078 | 1671.0 | 7.67% | 1112.4 | 7.13% | motif file (matrix) |
pdf |
| 36 |  | EBNA1(EBV virus)/Raji-EBNA1-ChIP-Seq(GSE30709)/Homer | 1e-2 | -6.817e+00 | 0.0078 | 33.0 | 0.15% | 13.4 | 0.09% | motif file (matrix) |
pdf |
| 37 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-2 | -6.766e+00 | 0.0078 | 1333.0 | 6.12% | 879.4 | 5.63% | motif file (matrix) |
pdf |
| 38 |  | Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer | 1e-2 | -6.647e+00 | 0.0084 | 678.0 | 3.11% | 432.4 | 2.77% | motif file (matrix) |
pdf |
| 39 | 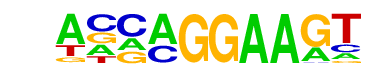 | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-2 | -6.400e+00 | 0.0105 | 2698.0 | 12.38% | 1831.7 | 11.74% | motif file (matrix) |
pdf |
| 40 |  | Bach1(bZIP)/K562-Bach1-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.854e+00 | 0.0176 | 117.0 | 0.54% | 64.2 | 0.41% | motif file (matrix) |
pdf |
| 41 |  | GATA-DR8(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-2 | -5.745e+00 | 0.0192 | 153.0 | 0.70% | 87.2 | 0.56% | motif file (matrix) |
pdf |
| 42 |  | STAT5(Stat)/mCD4+-Stat5-ChIP-Seq(GSE12346)/Homer | 1e-2 | -5.689e+00 | 0.0198 | 776.0 | 3.56% | 504.8 | 3.23% | motif file (matrix) |
pdf |
| 43 |  | PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-2 | -5.572e+00 | 0.0218 | 2822.0 | 12.95% | 1927.7 | 12.35% | motif file (matrix) |
pdf |
| 44 |  | CEBP:AP1(bZIP)/ThioMac-CEBPb-ChIP-Seq(GSE21512)/Homer | 1e-2 | -5.376e+00 | 0.0259 | 1574.0 | 7.22% | 1057.3 | 6.77% | motif file (matrix) |
pdf |
| 45 |  | CEBP:CEBP(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer | 1e-2 | -5.349e+00 | 0.0260 | 271.0 | 1.24% | 165.1 | 1.06% | motif file (matrix) |
pdf |
| 46 |  | USF1(HLH)/GM12878-Usf1-ChIP-Seq(GSE32465)/Homer | 1e-2 | -5.322e+00 | 0.0261 | 679.0 | 3.12% | 440.2 | 2.82% | motif file (matrix) |
pdf |
| 47 |  | T1ISRE(IRF)/Ifnb-Exp/Homer | 1e-2 | -5.187e+00 | 0.0292 | 15.0 | 0.07% | 5.9 | 0.04% | motif file (matrix) |
pdf |
| 48 |  | Mouse_Recombination_Hotspot/Testis-DMC1-ChIP-Seq(GSE24438)/Homer | 1e-2 | -5.117e+00 | 0.0307 | 138.0 | 0.63% | 80.0 | 0.51% | motif file (matrix) |
pdf |
| 49 |  | NFAT(RHD)/Jurkat-NFATC1-ChIP-Seq(Jolma et al.)/Homer | 1e-2 | -5.098e+00 | 0.0307 | 2022.0 | 9.28% | 1372.6 | 8.79% | motif file (matrix) |
pdf |
| 50 |  | E2A(HLH)/proBcell-E2A-ChIP-Seq(GSE21978)/Homer | 1e-2 | -4.796e+00 | 0.0406 | 2551.0 | 11.70% | 1746.5 | 11.19% | motif file (matrix) |
pdf |



































































































