| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | PU.1(ETS)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-8 | -1.998e+01 | 0.0000 | 154.0 | 4.56% | 56.5 | 2.77% | motif file (matrix) |
pdf |
| 2 |  | Olig2(bHLH)/Neuron-Olig2-ChIP-Seq(GSE30882)/Homer | 1e-7 | -1.808e+01 | 0.0000 | 744.0 | 22.02% | 372.1 | 18.22% | motif file (matrix) |
pdf |
| 3 |  | EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer | 1e-7 | -1.786e+01 | 0.0000 | 267.0 | 7.90% | 114.4 | 5.60% | motif file (matrix) |
pdf |
| 4 |  | ETS:RUNX/Jurkat-RUNX1-ChIP-Seq(GSE17954)/Homer | 1e-7 | -1.629e+01 | 0.0000 | 17.0 | 0.50% | 2.7 | 0.13% | motif file (matrix) |
pdf |
| 5 | 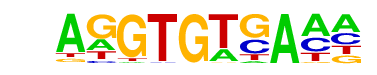 | Tbet(T-box)/CD8-Tbet-ChIP-Seq(GSE33802)/Homer | 1e-4 | -1.040e+01 | 0.0015 | 392.0 | 11.60% | 194.4 | 9.52% | motif file (matrix) |
pdf |
| 6 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-4 | -1.019e+01 | 0.0015 | 174.0 | 5.15% | 77.0 | 3.77% | motif file (matrix) |
pdf |
| 7 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-4 | -9.831e+00 | 0.0019 | 552.0 | 16.34% | 285.9 | 14.00% | motif file (matrix) |
pdf |
| 8 |  | Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer | 1e-4 | -9.751e+00 | 0.0019 | 110.0 | 3.26% | 45.5 | 2.23% | motif file (matrix) |
pdf |
| 9 |  | NFkB-p65(RHD)/GM12787-p65-ChIP-Seq(GSE19485)/Homer | 1e-3 | -9.127e+00 | 0.0030 | 140.0 | 4.14% | 61.3 | 3.00% | motif file (matrix) |
pdf |
| 10 |  | c-Myc(HLH)/LNCAP-cMyc-ChIP-Seq(unpublished)/Homer | 1e-3 | -8.922e+00 | 0.0033 | 104.0 | 3.08% | 43.8 | 2.15% | motif file (matrix) |
pdf |
| 11 |  | Maz(Zf)/HepG2-Maz-ChIP-Seq(GSE31477)/Homer | 1e-3 | -8.910e+00 | 0.0033 | 271.0 | 8.02% | 131.7 | 6.45% | motif file (matrix) |
pdf |
| 12 |  | NFY(CCAAT)/Promoter/Homer | 1e-3 | -8.876e+00 | 0.0033 | 189.0 | 5.59% | 87.4 | 4.28% | motif file (matrix) |
pdf |
| 13 |  | GATA3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-3 | -8.321e+00 | 0.0046 | 579.0 | 17.14% | 305.4 | 14.96% | motif file (matrix) |
pdf |
| 14 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-3 | -8.243e+00 | 0.0046 | 261.0 | 7.72% | 127.6 | 6.25% | motif file (matrix) |
pdf |
| 15 |  | Pitx1(Homeobox)/Chicken-Pitx1-ChIP-Seq(GSE38910)/Homer | 1e-3 | -8.070e+00 | 0.0051 | 1501.0 | 44.42% | 847.9 | 41.53% | motif file (matrix) |
pdf |
| 16 |  | GATA-IR4(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-3 | -7.761e+00 | 0.0065 | 39.0 | 1.15% | 13.2 | 0.65% | motif file (matrix) |
pdf |
| 17 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-3 | -7.656e+00 | 0.0068 | 331.0 | 9.80% | 167.7 | 8.21% | motif file (matrix) |
pdf |
| 18 |  | KLF5(Zf)/LoVo-KLF5-ChIP-Seq(GSE49402)/Homer | 1e-3 | -7.298e+00 | 0.0093 | 237.0 | 7.01% | 116.9 | 5.72% | motif file (matrix) |
pdf |
| 19 |  | TR4(NR/DR1)/Hela-TR4-ChIP-Seq(GSE24685)/Homer | 1e-3 | -6.971e+00 | 0.0122 | 31.0 | 0.92% | 10.4 | 0.51% | motif file (matrix) |
pdf |
| 20 |  | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-2 | -6.832e+00 | 0.0133 | 327.0 | 9.68% | 167.2 | 8.19% | motif file (matrix) |
pdf |
| 21 |  | NF-E2(bZIP)/K562-NFE2-ChIP-Seq(GSE31477)/Homer | 1e-2 | -6.585e+00 | 0.0162 | 16.0 | 0.47% | 4.7 | 0.23% | motif file (matrix) |
pdf |
| 22 |  | Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-2 | -6.451e+00 | 0.0176 | 74.0 | 2.19% | 31.0 | 1.52% | motif file (matrix) |
pdf |
| 23 |  | Eomes(T-box)/H9-Eomes-ChIP-Seq(GSE26097)/Homer | 1e-2 | -6.439e+00 | 0.0176 | 783.0 | 23.17% | 430.9 | 21.10% | motif file (matrix) |
pdf |
| 24 |  | Nrf2(bZIP)/Lymphoblast-Nrf2-ChIP-Seq(GSE37589)/Homer | 1e-2 | -6.271e+00 | 0.0194 | 13.0 | 0.38% | 3.4 | 0.17% | motif file (matrix) |
pdf |
| 25 |  | CEBP:CEBP(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer | 1e-2 | -6.228e+00 | 0.0194 | 45.0 | 1.33% | 18.0 | 0.88% | motif file (matrix) |
pdf |
| 26 |  | EKLF(Zf)/Erythrocyte-Klf1-ChIP-Seq(GSE20478)/Homer | 1e-2 | -6.086e+00 | 0.0215 | 53.0 | 1.57% | 21.6 | 1.06% | motif file (matrix) |
pdf |
| 27 |  | TATA-Box(TBP)/Promoter/Homer | 1e-2 | -6.074e+00 | 0.0215 | 471.0 | 13.94% | 251.1 | 12.30% | motif file (matrix) |
pdf |
| 28 |  | SCL(HLH)/HPC7-Scl-ChIP-Seq(GSE13511)/Homer | 1e-2 | -5.742e+00 | 0.0282 | 1536.0 | 45.46% | 880.9 | 43.14% | motif file (matrix) |
pdf |
| 29 |  | HIF-1a(HLH)/MCF7-HIF1a-ChIP-Seq(GSE28352)/Homer | 1e-2 | -5.642e+00 | 0.0301 | 68.0 | 2.01% | 30.0 | 1.47% | motif file (matrix) |
pdf |
| 30 |  | Bach2(bZIP)/OCILy7-Bach2-ChIP-Seq(GSE44420)/Homer | 1e-2 | -5.620e+00 | 0.0301 | 54.0 | 1.60% | 22.8 | 1.12% | motif file (matrix) |
pdf |
| 31 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-2 | -5.616e+00 | 0.0301 | 114.0 | 3.37% | 54.0 | 2.64% | motif file (matrix) |
pdf |
| 32 |  | Klf4(Zf)/mES-Klf4-ChIP-Seq(GSE11431)/Homer | 1e-2 | -5.461e+00 | 0.0327 | 85.0 | 2.52% | 38.5 | 1.89% | motif file (matrix) |
pdf |
| 33 |  | Arnt:Ahr(bHLH)/MCF7-Arnt-ChIP-Seq(Lo et al.)/Homer | 1e-2 | -5.418e+00 | 0.0331 | 156.0 | 4.62% | 77.0 | 3.77% | motif file (matrix) |
pdf |
| 34 |  | ETS:E-box/HPC7-Scl-ChIP-Seq(GSE22178)/Homer | 1e-2 | -5.404e+00 | 0.0331 | 35.0 | 1.04% | 13.4 | 0.66% | motif file (matrix) |
pdf |
| 35 | 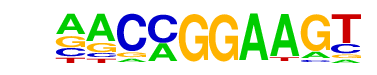 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-2 | -5.283e+00 | 0.0357 | 424.0 | 12.55% | 227.1 | 11.12% | motif file (matrix) |
pdf |
| 36 |  | Atoh1(bHLH)/Cerebellum-Atoh1-ChIP-Seq(GSE22111)/Homer | 1e-2 | -5.199e+00 | 0.0377 | 378.0 | 11.19% | 201.0 | 9.85% | motif file (matrix) |
pdf |
| 37 |  | PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-2 | -5.108e+00 | 0.0402 | 442.0 | 13.08% | 238.2 | 11.67% | motif file (matrix) |
pdf |
| 38 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-2 | -5.051e+00 | 0.0415 | 225.0 | 6.66% | 115.9 | 5.68% | motif file (matrix) |
pdf |
| 39 |  | FOXA1(Forkhead)/MCF7-FOXA1-ChIP-Seq(GSE26831)/Homer | 1e-2 | -4.967e+00 | 0.0439 | 352.0 | 10.42% | 187.7 | 9.19% | motif file (matrix) |
pdf |
| 40 |  | Tcf12(HLH)/GM12878-Tcf12-ChIP-Seq(GSE32465)/Homer | 1e-2 | -4.741e+00 | 0.0537 | 282.0 | 8.35% | 149.0 | 7.30% | motif file (matrix) |
pdf |
| 41 |  | NF1:FOXA1/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-2 | -4.687e+00 | 0.0553 | 23.0 | 0.68% | 8.4 | 0.41% | motif file (matrix) |
pdf |
| 42 |  | TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer | 1e-2 | -4.632e+00 | 0.0570 | 269.0 | 7.96% | 141.6 | 6.94% | motif file (matrix) |
pdf |



















































































