| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | CTCF(Zf)/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-768 | -1.770e+03 | 0.0000 | 1467.0 | 8.19% | 338.3 | 1.07% | motif file (matrix) |
pdf |
| 2 |  | BORIS(Zf)/K562-CTCFL-ChIP-Seq(GSE32465)/Homer | 1e-683 | -1.575e+03 | 0.0000 | 1674.0 | 9.35% | 528.5 | 1.66% | motif file (matrix) |
pdf |
| 3 |  | FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-676 | -1.557e+03 | 0.0000 | 4001.0 | 22.34% | 2734.4 | 8.61% | motif file (matrix) |
pdf |
| 4 |  | FOXA1(Forkhead)/MCF7-FOXA1-ChIP-Seq(GSE26831)/Homer | 1e-658 | -1.516e+03 | 0.0000 | 3406.0 | 19.02% | 2118.6 | 6.67% | motif file (matrix) |
pdf |
| 5 |  | Foxa2(Forkhead)/Liver-Foxa2-ChIP-Seq(GSE25694)/Homer | 1e-548 | -1.263e+03 | 0.0000 | 3046.0 | 17.01% | 1966.1 | 6.19% | motif file (matrix) |
pdf |
| 6 |  | Fox:Ebox(Forkhead:HLH)/Panc1-Foxa2-ChIP-Seq(GSE47459)/Homer | 1e-390 | -8.989e+02 | 0.0000 | 3540.0 | 19.77% | 2973.7 | 9.36% | motif file (matrix) |
pdf |
| 7 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-275 | -6.335e+02 | 0.0000 | 1538.0 | 8.59% | 969.4 | 3.05% | motif file (matrix) |
pdf |
| 8 |  | Unknown-ESC-element/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-231 | -5.333e+02 | 0.0000 | 2400.0 | 13.40% | 2094.3 | 6.59% | motif file (matrix) |
pdf |
| 9 |  | Foxo1(Forkhead)/RAW-Foxo1-ChIP-Seq(Fan et al.)/Homer | 1e-189 | -4.356e+02 | 0.0000 | 4662.0 | 26.03% | 5483.8 | 17.26% | motif file (matrix) |
pdf |
| 10 |  | Eomes(T-box)/H9-Eomes-ChIP-Seq(GSE26097)/Homer | 1e-172 | -3.979e+02 | 0.0000 | 4944.0 | 27.61% | 6027.8 | 18.97% | motif file (matrix) |
pdf |
| 11 |  | Lhx2(Homeobox)/HFSC-Lhx2-ChIP-Seq(GSE48068)/Homer | 1e-150 | -3.454e+02 | 0.0000 | 2439.0 | 13.62% | 2499.8 | 7.87% | motif file (matrix) |
pdf |
| 12 |  | Lhx3(Homeobox)/Neuron-Lhx3-ChIP-Seq(GSE31456)/Homer | 1e-142 | -3.276e+02 | 0.0000 | 3442.0 | 19.22% | 3975.4 | 12.51% | motif file (matrix) |
pdf |
| 13 |  | Gata4(Zf)/Heart-Gata4-ChIP-Seq(GSE35151)/Homer | 1e-133 | -3.078e+02 | 0.0000 | 2361.0 | 13.18% | 2478.6 | 7.80% | motif file (matrix) |
pdf |
| 14 |  | Gata2(Zf)/K562-GATA2-ChIP-Seq(GSE18829)/Homer | 1e-132 | -3.048e+02 | 0.0000 | 1688.0 | 9.43% | 1580.9 | 4.98% | motif file (matrix) |
pdf |
| 15 |  | GATA3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-125 | -2.884e+02 | 0.0000 | 3172.0 | 17.71% | 3692.5 | 11.62% | motif file (matrix) |
pdf |
| 16 |  | Gata1(Zf)/K562-GATA1-ChIP-Seq(GSE18829)/Homer | 1e-120 | -2.784e+02 | 0.0000 | 1532.0 | 8.56% | 1428.9 | 4.50% | motif file (matrix) |
pdf |
| 17 |  | Tbx5(T-box)/HL1-Tbx5.biotin-ChIP-Seq(GSE21529)/Homer | 1e-108 | -2.501e+02 | 0.0000 | 7416.0 | 41.41% | 10629.4 | 33.46% | motif file (matrix) |
pdf |
| 18 |  | Nkx6.1(Homeobox)/Islet-Nkx6.1-ChIP-Seq(GSE40975)/Homer | 1e-105 | -2.422e+02 | 0.0000 | 4780.0 | 26.69% | 6331.4 | 19.93% | motif file (matrix) |
pdf |
| 19 |  | Tbet(T-box)/CD8-Tbet-ChIP-Seq(GSE33802)/Homer | 1e-100 | -2.319e+02 | 0.0000 | 2519.0 | 14.07% | 2904.8 | 9.14% | motif file (matrix) |
pdf |
| 20 |  | Sox3(HMG)/NPC-Sox3-ChIP-Seq(GSE33059)/Homer | 1e-91 | -2.111e+02 | 0.0000 | 3759.0 | 20.99% | 4850.9 | 15.27% | motif file (matrix) |
pdf |
| 21 |  | Sox2(HMG)/mES-Sox2-ChIP-Seq(GSE11431)/Homer | 1e-79 | -1.839e+02 | 0.0000 | 2040.0 | 11.39% | 2353.2 | 7.41% | motif file (matrix) |
pdf |
| 22 |  | Sox6(HMG)/Myotubes-Sox6-ChIP-Seq(GSE32627)/Homer | 1e-76 | -1.767e+02 | 0.0000 | 3261.0 | 18.21% | 4216.7 | 13.27% | motif file (matrix) |
pdf |
| 23 |  | Nanog(Homeobox)/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-72 | -1.668e+02 | 0.0000 | 7914.0 | 44.20% | 11943.1 | 37.59% | motif file (matrix) |
pdf |
| 24 |  | CRX(Homeobox)/Retina-Crx-ChIP-Seq(GSE20012)/Homer | 1e-68 | -1.577e+02 | 0.0000 | 4684.0 | 26.16% | 6572.9 | 20.69% | motif file (matrix) |
pdf |
| 25 |  | Oct4(POU/Homeobox)/mES-Oct4-ChIP-Seq(GSE11431)/Homer | 1e-64 | -1.489e+02 | 0.0000 | 881.0 | 4.92% | 838.3 | 2.64% | motif file (matrix) |
pdf |
| 26 |  | Ptf1a(HLH)/Panc1-Ptf1a-ChIP-Seq(GSE47459)/Homer | 1e-58 | -1.343e+02 | 0.0000 | 6123.0 | 34.19% | 9097.9 | 28.64% | motif file (matrix) |
pdf |
| 27 |  | SCL(HLH)/HPC7-Scl-ChIP-Seq(GSE13511)/Homer | 1e-57 | -1.319e+02 | 0.0000 | 9804.0 | 54.75% | 15493.2 | 48.77% | motif file (matrix) |
pdf |
| 28 |  | Isl1(Homeobox)/Neuron-Isl1-ChIP-Seq(GSE31456)/Homer | 1e-54 | -1.259e+02 | 0.0000 | 3996.0 | 22.32% | 5627.3 | 17.71% | motif file (matrix) |
pdf |
| 29 |  | E2A(HLH)/proBcell-E2A-ChIP-Seq(GSE21978)/Homer | 1e-50 | -1.173e+02 | 0.0000 | 3727.0 | 20.81% | 5240.1 | 16.49% | motif file (matrix) |
pdf |
| 30 |  | Tcf4(HMG)/Hct116-Tcf4-ChIP-Seq(SRA012054)/Homer | 1e-49 | -1.141e+02 | 0.0000 | 1018.0 | 5.68% | 1102.5 | 3.47% | motif file (matrix) |
pdf |
| 31 |  | Tcf3(HMG)/mES-Tcf3-ChIP-Seq(GSE11724)/Homer | 1e-45 | -1.052e+02 | 0.0000 | 635.0 | 3.55% | 608.6 | 1.92% | motif file (matrix) |
pdf |
| 32 |  | REST-NRSF(Zf)/Jurkat-NRSF-ChIP-Seq/Homer | 1e-42 | -9.671e+01 | 0.0000 | 81.0 | 0.45% | 19.8 | 0.06% | motif file (matrix) |
pdf |
| 33 |  | Tcf12(HLH)/GM12878-Tcf12-ChIP-Seq(GSE32465)/Homer | 1e-41 | -9.670e+01 | 0.0000 | 2631.0 | 14.69% | 3599.3 | 11.33% | motif file (matrix) |
pdf |
| 34 |  | E2A-nearPU.1(HLH)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-38 | -8.833e+01 | 0.0000 | 3391.0 | 18.94% | 4868.8 | 15.33% | motif file (matrix) |
pdf |
| 35 |  | Phox2a(Homeobox)/Neuron-Phox2a-ChIP-Seq(GSE31456)/Homer | 1e-37 | -8.536e+01 | 0.0000 | 616.0 | 3.44% | 626.8 | 1.97% | motif file (matrix) |
pdf |
| 36 |  | Oct2(POU/Homeobox)/Bcell-Oct2-ChIP-Seq(GSE21512)/Homer | 1e-36 | -8.499e+01 | 0.0000 | 494.0 | 2.76% | 467.9 | 1.47% | motif file (matrix) |
pdf |
| 37 |  | KLF5(Zf)/LoVo-KLF5-ChIP-Seq(GSE49402)/Homer | 1e-36 | -8.458e+01 | 0.0000 | 2989.0 | 16.69% | 4241.0 | 13.35% | motif file (matrix) |
pdf |
| 38 |  | Pitx1(Homeobox)/Chicken-Pitx1-ChIP-Seq(GSE38910)/Homer | 1e-36 | -8.428e+01 | 0.0000 | 7552.0 | 42.17% | 11925.8 | 37.54% | motif file (matrix) |
pdf |
| 39 |  | Ap4(HLH)/AML-Tfap4-ChIP-Seq(GSE45738)/Homer | 1e-32 | -7.549e+01 | 0.0000 | 2950.0 | 16.47% | 4234.7 | 13.33% | motif file (matrix) |
pdf |
| 40 |  | NeuroD1(bHLH)/Islet-NeuroD1-ChIP-Seq(GSE30298)/Homer | 1e-32 | -7.545e+01 | 0.0000 | 1907.0 | 10.65% | 2571.7 | 8.09% | motif file (matrix) |
pdf |
| 41 |  | OCT4-SOX2-TCF-NANOG((POU/Homeobox/HMG)/mES-Oct4-ChIP-Seq(GSE11431)/Homer | 1e-32 | -7.457e+01 | 0.0000 | 287.0 | 1.60% | 231.0 | 0.73% | motif file (matrix) |
pdf |
| 42 |  | RFX(HTH)/K562-RFX3-ChIP-Seq(SRA012198)/Homer | 1e-31 | -7.192e+01 | 0.0000 | 276.0 | 1.54% | 221.6 | 0.70% | motif file (matrix) |
pdf |
| 43 |  | TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer | 1e-29 | -6.698e+01 | 0.0000 | 1622.0 | 9.06% | 2170.8 | 6.83% | motif file (matrix) |
pdf |
| 44 |  | CTCF-SatelliteElement/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-28 | -6.473e+01 | 0.0000 | 83.0 | 0.46% | 32.4 | 0.10% | motif file (matrix) |
pdf |
| 45 |  | TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer | 1e-27 | -6.337e+01 | 0.0000 | 1210.0 | 6.76% | 1554.6 | 4.89% | motif file (matrix) |
pdf |
| 46 |  | Rfx2(HTH)/LoVo-RFX2-ChIP-Seq(GSE49402)/Homer | 1e-25 | -5.933e+01 | 0.0000 | 307.0 | 1.71% | 279.4 | 0.88% | motif file (matrix) |
pdf |
| 47 |  | Foxh1(Forkhead)/hESC-FOXH1-ChIP-Seq(GSE29422)/Homer | 1e-25 | -5.793e+01 | 0.0000 | 1043.0 | 5.82% | 1325.8 | 4.17% | motif file (matrix) |
pdf |
| 48 |  | HNF6(Homeobox)/Liver-Hnf6-ChIP-Seq(ERP000394)/Homer | 1e-24 | -5.685e+01 | 0.0000 | 807.0 | 4.51% | 978.5 | 3.08% | motif file (matrix) |
pdf |
| 49 |  | Maz(Zf)/HepG2-Maz-ChIP-Seq(GSE31477)/Homer | 1e-24 | -5.615e+01 | 0.0000 | 3186.0 | 17.79% | 4759.8 | 14.98% | motif file (matrix) |
pdf |
| 50 |  | Sp1(Zf)/Promoter/Homer | 1e-24 | -5.592e+01 | 0.0000 | 602.0 | 3.36% | 685.7 | 2.16% | motif file (matrix) |
pdf |
| 51 |  | EBF1(EBF)/Near-E2A-ChIP-Seq(GSE21512)/Homer | 1e-23 | -5.431e+01 | 0.0000 | 2471.0 | 13.80% | 3600.9 | 11.33% | motif file (matrix) |
pdf |
| 52 |  | MyoG(HLH)/C2C12-MyoG-ChIP-Seq(GSE36024)/Homer | 1e-22 | -5.266e+01 | 0.0000 | 2585.0 | 14.44% | 3798.3 | 11.96% | motif file (matrix) |
pdf |
| 53 |  | X-box(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-21 | -4.895e+01 | 0.0000 | 312.0 | 1.74% | 306.0 | 0.96% | motif file (matrix) |
pdf |
| 54 |  | TCFL2(HMG)/K562-TCF7L2-ChIP-Seq(GSE29196)/Homer | 1e-18 | -4.161e+01 | 0.0000 | 225.0 | 1.26% | 209.0 | 0.66% | motif file (matrix) |
pdf |
| 55 |  | Olig2(bHLH)/Neuron-Olig2-ChIP-Seq(GSE30882)/Homer | 1e-17 | -4.106e+01 | 0.0000 | 3863.0 | 21.57% | 6028.3 | 18.98% | motif file (matrix) |
pdf |
| 56 |  | Myf5(bHLH)/GM-Myf5-ChIP-Seq(GSE24852)/Homer | 1e-17 | -4.074e+01 | 0.0000 | 1730.0 | 9.66% | 2496.6 | 7.86% | motif file (matrix) |
pdf |
| 57 |  | Rfx1(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-16 | -3.799e+01 | 0.0000 | 494.0 | 2.76% | 589.2 | 1.85% | motif file (matrix) |
pdf |
| 58 |  | Rfx5(HTH)/GM12878-Rfx5-ChIP-Seq(GSE31477)/Homer | 1e-16 | -3.749e+01 | 0.0000 | 711.0 | 3.97% | 913.4 | 2.88% | motif file (matrix) |
pdf |
| 59 |  | Atoh1(bHLH)/Cerebellum-Atoh1-ChIP-Seq(GSE22111)/Homer | 1e-15 | -3.595e+01 | 0.0000 | 2402.0 | 13.41% | 3633.5 | 11.44% | motif file (matrix) |
pdf |
| 60 |  | FOXA1:AR/LNCAP-AR-ChIP-Seq(GSE27824)/Homer | 1e-15 | -3.543e+01 | 0.0000 | 120.0 | 0.67% | 92.7 | 0.29% | motif file (matrix) |
pdf |
| 61 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-14 | -3.242e+01 | 0.0000 | 3258.0 | 18.19% | 5099.3 | 16.05% | motif file (matrix) |
pdf |
| 62 |  | Oct4:Sox17/F9-Sox17-ChIP-Seq(GSE44553)/Homer | 1e-13 | -3.014e+01 | 0.0000 | 256.0 | 1.43% | 276.6 | 0.87% | motif file (matrix) |
pdf |
| 63 |  | AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-12 | -2.977e+01 | 0.0000 | 1212.0 | 6.77% | 1740.9 | 5.48% | motif file (matrix) |
pdf |
| 64 |  | Tbox:Smad/ESCd5-Smad2_3-ChIP-Seq(GSE29422)/Homer | 1e-12 | -2.973e+01 | 0.0000 | 460.0 | 2.57% | 570.9 | 1.80% | motif file (matrix) |
pdf |
| 65 |  | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-12 | -2.857e+01 | 0.0000 | 2782.0 | 15.54% | 4340.1 | 13.66% | motif file (matrix) |
pdf |
| 66 |  | MafA(bZIP)/Islet-MafA-ChIP-Seq(GSE30298)/Homer | 1e-12 | -2.854e+01 | 0.0000 | 1713.0 | 9.57% | 2563.2 | 8.07% | motif file (matrix) |
pdf |
| 67 |  | Tbx20(T-box)/Heart-Tbx20-ChIP-Seq(GSE29636)/Homer | 1e-11 | -2.751e+01 | 0.0000 | 496.0 | 2.77% | 633.4 | 1.99% | motif file (matrix) |
pdf |
| 68 |  | EBF(EBF)/proBcell-EBF-ChIP-Seq(GSE21978)/Homer | 1e-11 | -2.692e+01 | 0.0000 | 530.0 | 2.96% | 687.4 | 2.16% | motif file (matrix) |
pdf |
| 69 |  | Rbpj1(?)/Panc1-Rbpj1-ChIP-Seq(GSE47459)/Homer | 1e-11 | -2.674e+01 | 0.0000 | 2815.0 | 15.72% | 4417.0 | 13.90% | motif file (matrix) |
pdf |
| 70 |  | Hnf1(Homeobox)/Liver-Foxa2-Chip-Seq(GSE25694)/Homer | 1e-11 | -2.593e+01 | 0.0000 | 184.0 | 1.03% | 189.6 | 0.60% | motif file (matrix) |
pdf |
| 71 |  | Pax7-long(Paired/Homeobox)/Myoblast-Pax7-ChIP-Seq(GSE25064)/Homer | 1e-11 | -2.577e+01 | 0.0000 | 57.0 | 0.32% | 35.7 | 0.11% | motif file (matrix) |
pdf |
| 72 |  | PU.1(ETS)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-10 | -2.500e+01 | 0.0000 | 952.0 | 5.32% | 1357.4 | 4.27% | motif file (matrix) |
pdf |
| 73 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-10 | -2.500e+01 | 0.0000 | 1032.0 | 5.76% | 1485.7 | 4.68% | motif file (matrix) |
pdf |
| 74 |  | NF1:FOXA1/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-10 | -2.449e+01 | 0.0000 | 116.0 | 0.65% | 104.9 | 0.33% | motif file (matrix) |
pdf |
| 75 |  | Pax7(Paired/Homeobox)/Myoblast-Pax7-ChIP-Seq(GSE25064)/Homer | 1e-10 | -2.358e+01 | 0.0000 | 125.0 | 0.70% | 117.8 | 0.37% | motif file (matrix) |
pdf |
| 76 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-9 | -2.265e+01 | 0.0000 | 1043.0 | 5.82% | 1520.8 | 4.79% | motif file (matrix) |
pdf |
| 77 |  | Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-9 | -2.152e+01 | 0.0000 | 400.0 | 2.23% | 515.1 | 1.62% | motif file (matrix) |
pdf |
| 78 |  | SPDEF(ETS)/VCaP-SPDEF-ChIP-Seq(SRA014231)/Homer | 1e-9 | -2.102e+01 | 0.0000 | 2004.0 | 11.19% | 3119.5 | 9.82% | motif file (matrix) |
pdf |
| 79 |  | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-8 | -2.055e+01 | 0.0000 | 2135.0 | 11.92% | 3345.0 | 10.53% | motif file (matrix) |
pdf |
| 80 |  | c-Jun-CRE(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-8 | -2.012e+01 | 0.0000 | 502.0 | 2.80% | 678.8 | 2.14% | motif file (matrix) |
pdf |
| 81 |  | BMYB(HTH)/Hela-BMYB-ChIPSeq(GSE27030)/Homer | 1e-8 | -1.996e+01 | 0.0000 | 2713.0 | 15.15% | 4327.7 | 13.62% | motif file (matrix) |
pdf |
| 82 |  | Egr1(Zf)/K562-Egr1-ChIP-Seq(GSE32465)/Homer | 1e-8 | -1.990e+01 | 0.0000 | 1311.0 | 7.32% | 1980.7 | 6.23% | motif file (matrix) |
pdf |
| 83 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-8 | -1.911e+01 | 0.0000 | 2010.0 | 11.22% | 3151.6 | 9.92% | motif file (matrix) |
pdf |
| 84 |  | Hoxb4(Homeobox)/ES-Hoxb4-ChIP-Seq(GSE34014)/Homer | 1e-7 | -1.838e+01 | 0.0000 | 297.0 | 1.66% | 375.0 | 1.18% | motif file (matrix) |
pdf |
| 85 |  | Erra(NR)/HepG2-Erra-ChIP-Seq(GSE31477)/Homer | 1e-7 | -1.816e+01 | 0.0000 | 4246.0 | 23.71% | 6980.1 | 21.97% | motif file (matrix) |
pdf |
| 86 |  | Klf4(Zf)/mES-Klf4-ChIP-Seq(GSE11431)/Homer | 1e-7 | -1.799e+01 | 0.0000 | 804.0 | 4.49% | 1170.2 | 3.68% | motif file (matrix) |
pdf |
| 87 |  | Bach2(bZIP)/OCILy7-Bach2-ChIP-Seq(GSE44420)/Homer | 1e-7 | -1.699e+01 | 0.0000 | 357.0 | 1.99% | 471.3 | 1.48% | motif file (matrix) |
pdf |
| 88 |  | GATA:SCL/Ter119-SCL-ChIP-Seq(GSE18720)/Homer | 1e-7 | -1.661e+01 | 0.0000 | 243.0 | 1.36% | 301.6 | 0.95% | motif file (matrix) |
pdf |
| 89 |  | Atf1(bZIP)/K562-ATF1-ChIP-Seq(GSE31477)/Homer | 1e-7 | -1.619e+01 | 0.0000 | 985.0 | 5.50% | 1479.7 | 4.66% | motif file (matrix) |
pdf |
| 90 |  | EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer | 1e-6 | -1.610e+01 | 0.0000 | 1298.0 | 7.25% | 1996.5 | 6.28% | motif file (matrix) |
pdf |
| 91 |  | MyoD(HLH)/Myotube-MyoD-ChIP-Seq(GSE21614)/Homer | 1e-6 | -1.594e+01 | 0.0000 | 1805.0 | 10.08% | 2845.2 | 8.96% | motif file (matrix) |
pdf |
| 92 |  | NFY(CCAAT)/Promoter/Homer | 1e-6 | -1.594e+01 | 0.0000 | 1371.0 | 7.66% | 2119.4 | 6.67% | motif file (matrix) |
pdf |
| 93 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-6 | -1.592e+01 | 0.0000 | 1124.0 | 6.28% | 1710.9 | 5.39% | motif file (matrix) |
pdf |
| 94 |  | AMYB(HTH)/Testes-AMYB-ChIP-Seq(GSE44588)/Homer | 1e-6 | -1.573e+01 | 0.0000 | 2795.0 | 15.61% | 4527.8 | 14.25% | motif file (matrix) |
pdf |
| 95 |  | Znf263(Zf)/K562-Znf263-ChIP-Seq(GSE31477)/Homer | 1e-6 | -1.556e+01 | 0.0000 | 3878.0 | 21.66% | 6390.6 | 20.12% | motif file (matrix) |
pdf |
| 96 |  | MafK(bZIP)/C2C12-MafK-ChIP-Seq(GSE36030)/Homer | 1e-6 | -1.507e+01 | 0.0000 | 452.0 | 2.52% | 629.4 | 1.98% | motif file (matrix) |
pdf |
| 97 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-5 | -1.354e+01 | 0.0000 | 1758.0 | 9.82% | 2797.4 | 8.81% | motif file (matrix) |
pdf |
| 98 |  | E2F6(E2F)/Hela-E2F6-ChIP-Seq(GSE31477)/Homer | 1e-5 | -1.345e+01 | 0.0000 | 833.0 | 4.65% | 1255.7 | 3.95% | motif file (matrix) |
pdf |
| 99 |  | Srebp2(HLH)/HepG2-Srebp2-ChIP-Seq(GSE31477)/Homer | 1e-5 | -1.309e+01 | 0.0000 | 264.0 | 1.47% | 348.5 | 1.10% | motif file (matrix) |
pdf |
| 100 |  | PAX3:FKHR-fusion(Paired/Homeobox)/Rh4-PAX3:FKHR-ChIP-Seq(GSE19063)/Homer | 1e-5 | -1.255e+01 | 0.0000 | 296.0 | 1.65% | 401.0 | 1.26% | motif file (matrix) |
pdf |
| 101 |  | Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer | 1e-5 | -1.188e+01 | 0.0000 | 639.0 | 3.57% | 952.9 | 3.00% | motif file (matrix) |
pdf |
| 102 | 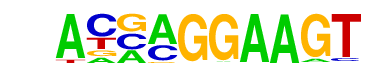 | ELF5(ETS)/T47D-ELF5-ChIP-Seq(GSE30407)/Homer | 1e-4 | -1.104e+01 | 0.0000 | 1377.0 | 7.69% | 2188.1 | 6.89% | motif file (matrix) |
pdf |
| 103 |  | CRE(bZIP)/Promoter/Homer | 1e-4 | -1.100e+01 | 0.0000 | 335.0 | 1.87% | 470.2 | 1.48% | motif file (matrix) |
pdf |
| 104 |  | Nur77(NR)/K562-NR4A1-ChIP-Seq(GSE31363)/Homer | 1e-4 | -1.099e+01 | 0.0000 | 300.0 | 1.68% | 415.5 | 1.31% | motif file (matrix) |
pdf |
| 105 |  | Srebp1a(HLH)/HepG2-Srebp1a-ChIP-Seq(GSE31477)/Homer | 1e-4 | -1.033e+01 | 0.0001 | 365.0 | 2.04% | 522.2 | 1.64% | motif file (matrix) |
pdf |
| 106 |  | NF1-halfsite(CTF)/LNCaP-NF1-ChIP-Seq(Unpublished)/Homer | 1e-4 | -9.601e+00 | 0.0002 | 3465.0 | 19.35% | 5794.4 | 18.24% | motif file (matrix) |
pdf |
| 107 |  | PAX5-shortForm(Paired/Homeobox)/GM12878-PAX5-ChIP-Seq(GSE32465)/Homer | 1e-4 | -9.492e+00 | 0.0002 | 189.0 | 1.06% | 251.2 | 0.79% | motif file (matrix) |
pdf |
| 108 |  | Bcl6(Zf)/Liver-Bcl6-ChIP-Seq(GSE31578)/Homer | 1e-4 | -9.346e+00 | 0.0002 | 2321.0 | 12.96% | 3824.9 | 12.04% | motif file (matrix) |
pdf |
| 109 |  | Nkx3.1(Homeobox)/LNCaP-Nkx3.1-ChIP-Seq(GSE28264)/Homer | 1e-4 | -9.340e+00 | 0.0002 | 4071.0 | 22.73% | 6853.5 | 21.57% | motif file (matrix) |
pdf |
| 110 |  | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-4 | -9.215e+00 | 0.0002 | 2362.0 | 13.19% | 3897.6 | 12.27% | motif file (matrix) |
pdf |
| 111 |  | RUNX-AML(Runt)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer | 1e-3 | -9.021e+00 | 0.0003 | 1334.0 | 7.45% | 2144.3 | 6.75% | motif file (matrix) |
pdf |
| 112 |  | GATA-IR4(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-3 | -8.992e+00 | 0.0003 | 124.0 | 0.69% | 155.2 | 0.49% | motif file (matrix) |
pdf |
| 113 |  | Pdx1(Homeobox)/Islet-Pdx1-ChIP-Seq(SRA008281)/Homer | 1e-3 | -8.648e+00 | 0.0004 | 1300.0 | 7.26% | 2092.1 | 6.59% | motif file (matrix) |
pdf |
| 114 |  | RBPJ:Ebox/Panc1-Rbpj1-ChIP-Seq(GSE47459)/Homer | 1e-3 | -8.526e+00 | 0.0004 | 633.0 | 3.53% | 974.7 | 3.07% | motif file (matrix) |
pdf |
| 115 |  | JunD(bZIP)/K562-JunD-ChIP-Seq/Homer | 1e-3 | -8.283e+00 | 0.0005 | 159.0 | 0.89% | 211.4 | 0.67% | motif file (matrix) |
pdf |
| 116 |  | Nrf2(bZIP)/Lymphoblast-Nrf2-ChIP-Seq(GSE37589)/Homer | 1e-3 | -8.018e+00 | 0.0007 | 82.0 | 0.46% | 97.1 | 0.31% | motif file (matrix) |
pdf |
| 117 |  | bZIP:IRF/Th17-BatF-ChIP-Seq(GSE39756)/Homer | 1e-3 | -7.985e+00 | 0.0007 | 554.0 | 3.09% | 849.8 | 2.67% | motif file (matrix) |
pdf |
| 118 |  | NF-E2(bZIP)/K562-NFE2-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.950e+00 | 0.0007 | 101.0 | 0.56% | 125.4 | 0.39% | motif file (matrix) |
pdf |
| 119 |  | Six1(Homeobox)/Myoblast-Six1-ChIP-Chip(GSE20150)/Homer | 1e-3 | -7.923e+00 | 0.0007 | 373.0 | 2.08% | 553.9 | 1.74% | motif file (matrix) |
pdf |
| 120 |  | E2F4(E2F)/K562-E2F4-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.446e+00 | 0.0012 | 594.0 | 3.32% | 921.7 | 2.90% | motif file (matrix) |
pdf |
| 121 |  | PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-3 | -7.349e+00 | 0.0013 | 2168.0 | 12.11% | 3601.0 | 11.33% | motif file (matrix) |
pdf |
| 122 |  | Usf2(HLH)/C2C12-Usf2-ChIP-Seq(GSE36030)/Homer | 1e-3 | -7.282e+00 | 0.0014 | 480.0 | 2.68% | 734.3 | 2.31% | motif file (matrix) |
pdf |
| 123 |  | NFkB-p65(RHD)/GM12787-p65-ChIP-Seq(GSE19485)/Homer | 1e-3 | -7.200e+00 | 0.0015 | 880.0 | 4.91% | 1403.6 | 4.42% | motif file (matrix) |
pdf |
| 124 |  | GATA-DR4(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-3 | -7.079e+00 | 0.0017 | 108.0 | 0.60% | 139.2 | 0.44% | motif file (matrix) |
pdf |
| 125 |  | MYB(HTH)/ERMYB-Myb-ChIPSeq(GSE22095)/Homer | 1e-2 | -6.809e+00 | 0.0022 | 3000.0 | 16.75% | 5054.6 | 15.91% | motif file (matrix) |
pdf |
| 126 |  | Gfi1b(Zf)/HPC7-Gfi1b-ChIP-Seq(GSE22178)/Homer | 1e-2 | -6.214e+00 | 0.0039 | 1040.0 | 5.81% | 1689.2 | 5.32% | motif file (matrix) |
pdf |
| 127 |  | CEBP:AP1(bZIP)/ThioMac-CEBPb-ChIP-Seq(GSE21512)/Homer | 1e-2 | -5.837e+00 | 0.0057 | 1048.0 | 5.85% | 1709.6 | 5.38% | motif file (matrix) |
pdf |
| 128 |  | GATA-DR8(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-2 | -5.398e+00 | 0.0087 | 97.0 | 0.54% | 130.2 | 0.41% | motif file (matrix) |
pdf |
| 129 |  | Unknown(Homeobox)/Limb-p300-ChIP-Seq/Homer | 1e-2 | -5.341e+00 | 0.0091 | 844.0 | 4.71% | 1370.4 | 4.31% | motif file (matrix) |
pdf |
| 130 |  | PRDM1/BMI1(Zf)/Hela-PRDM1-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.209e+00 | 0.0103 | 778.0 | 4.34% | 1260.4 | 3.97% | motif file (matrix) |
pdf |
| 131 |  | EKLF(Zf)/Erythrocyte-Klf1-ChIP-Seq(GSE20478)/Homer | 1e-2 | -5.109e+00 | 0.0113 | 444.0 | 2.48% | 698.8 | 2.20% | motif file (matrix) |
pdf |
| 132 |  | TATA-Box(TBP)/Promoter/Homer | 1e-2 | -4.804e+00 | 0.0153 | 1954.0 | 10.91% | 3291.1 | 10.36% | motif file (matrix) |
pdf |
| 133 |  | PBX1(Homeobox)/MCF7-PBX1-ChIP-Seq(GSE28007)/Homer | 1e-2 | -4.788e+00 | 0.0154 | 168.0 | 0.94% | 246.7 | 0.78% | motif file (matrix) |
pdf |
| 134 |  | Max(HLH)/K562-Max-ChIP-Seq(GSE31477)/Homer | 1e-2 | -4.715e+00 | 0.0164 | 932.0 | 5.20% | 1531.1 | 4.82% | motif file (matrix) |
pdf |
| 135 |  | ETS:E-box/HPC7-Scl-ChIP-Seq(GSE22178)/Homer | 1e-2 | -4.704e+00 | 0.0165 | 188.0 | 1.05% | 279.3 | 0.88% | motif file (matrix) |
pdf |













































































































































































































































































