| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-298 | -6.877e+02 | 0.0000 | 2815.0 | 19.65% | 1175.6 | 9.46% | motif file (matrix) |
pdf |
| 2 |  | FOXA1(Forkhead)/MCF7-FOXA1-ChIP-Seq(GSE26831)/Homer | 1e-284 | -6.556e+02 | 0.0000 | 2383.0 | 16.64% | 935.7 | 7.53% | motif file (matrix) |
pdf |
| 3 |  | Foxa2(Forkhead)/Liver-Foxa2-ChIP-Seq(GSE25694)/Homer | 1e-269 | -6.213e+02 | 0.0000 | 2065.0 | 14.42% | 771.4 | 6.21% | motif file (matrix) |
pdf |
| 4 |  | Fox:Ebox(Forkhead:HLH)/Panc1-Foxa2-ChIP-Seq(GSE47459)/Homer | 1e-145 | -3.362e+02 | 0.0000 | 2391.0 | 16.69% | 1209.4 | 9.74% | motif file (matrix) |
pdf |
| 5 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-125 | -2.892e+02 | 0.0000 | 1044.0 | 7.29% | 398.1 | 3.20% | motif file (matrix) |
pdf |
| 6 |  | CTCF(Zf)/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-119 | -2.746e+02 | 0.0000 | 536.0 | 3.74% | 141.6 | 1.14% | motif file (matrix) |
pdf |
| 7 |  | BORIS(Zf)/K562-CTCFL-ChIP-Seq(GSE32465)/Homer | 1e-92 | -2.134e+02 | 0.0000 | 614.0 | 4.29% | 207.3 | 1.67% | motif file (matrix) |
pdf |
| 8 |  | Unknown-ESC-element/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-89 | -2.061e+02 | 0.0000 | 1488.0 | 10.39% | 746.8 | 6.01% | motif file (matrix) |
pdf |
| 9 |  | Lhx3(Homeobox)/Neuron-Lhx3-ChIP-Seq(GSE31456)/Homer | 1e-83 | -1.920e+02 | 0.0000 | 2644.0 | 18.46% | 1583.1 | 12.75% | motif file (matrix) |
pdf |
| 10 |  | Lhx2(Homeobox)/HFSC-Lhx2-ChIP-Seq(GSE48068)/Homer | 1e-82 | -1.901e+02 | 0.0000 | 1871.0 | 13.06% | 1028.6 | 8.28% | motif file (matrix) |
pdf |
| 11 |  | Foxo1(Forkhead)/RAW-Foxo1-ChIP-Seq(Fan et al.)/Homer | 1e-77 | -1.774e+02 | 0.0000 | 3294.0 | 22.99% | 2100.6 | 16.91% | motif file (matrix) |
pdf |
| 12 |  | Gata2(Zf)/K562-GATA2-ChIP-Seq(GSE18829)/Homer | 1e-76 | -1.762e+02 | 0.0000 | 1222.0 | 8.53% | 603.1 | 4.86% | motif file (matrix) |
pdf |
| 13 |  | Gata4(Zf)/Heart-Gata4-ChIP-Seq(GSE35151)/Homer | 1e-74 | -1.718e+02 | 0.0000 | 1725.0 | 12.04% | 951.9 | 7.66% | motif file (matrix) |
pdf |
| 14 |  | Gata1(Zf)/K562-GATA1-ChIP-Seq(GSE18829)/Homer | 1e-69 | -1.601e+02 | 0.0000 | 1084.0 | 7.57% | 530.5 | 4.27% | motif file (matrix) |
pdf |
| 15 |  | GATA3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-58 | -1.340e+02 | 0.0000 | 2407.0 | 16.80% | 1512.1 | 12.17% | motif file (matrix) |
pdf |
| 16 |  | Nkx6.1(Homeobox)/Islet-Nkx6.1-ChIP-Seq(GSE40975)/Homer | 1e-58 | -1.336e+02 | 0.0000 | 3795.0 | 26.49% | 2590.9 | 20.86% | motif file (matrix) |
pdf |
| 17 |  | Eomes(T-box)/H9-Eomes-ChIP-Seq(GSE26097)/Homer | 1e-53 | -1.232e+02 | 0.0000 | 3575.0 | 24.96% | 2442.2 | 19.66% | motif file (matrix) |
pdf |
| 18 |  | Isl1(Homeobox)/Neuron-Isl1-ChIP-Seq(GSE31456)/Homer | 1e-45 | -1.039e+02 | 0.0000 | 3014.0 | 21.04% | 2049.1 | 16.50% | motif file (matrix) |
pdf |
| 19 |  | Sox3(HMG)/NPC-Sox3-ChIP-Seq(GSE33059)/Homer | 1e-40 | -9.326e+01 | 0.0000 | 2767.0 | 19.32% | 1883.2 | 15.16% | motif file (matrix) |
pdf |
| 20 |  | Sox2(HMG)/mES-Sox2-ChIP-Seq(GSE11431)/Homer | 1e-34 | -7.889e+01 | 0.0000 | 1471.0 | 10.27% | 923.1 | 7.43% | motif file (matrix) |
pdf |
| 21 | 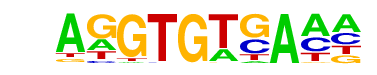 | Tbet(T-box)/CD8-Tbet-ChIP-Seq(GSE33802)/Homer | 1e-31 | -7.165e+01 | 0.0000 | 1862.0 | 13.00% | 1236.3 | 9.95% | motif file (matrix) |
pdf |
| 22 |  | Nanog(Homeobox)/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-30 | -7.014e+01 | 0.0000 | 5889.0 | 41.11% | 4524.3 | 36.42% | motif file (matrix) |
pdf |
| 23 |  | CRX(Homeobox)/Retina-Crx-ChIP-Seq(GSE20012)/Homer | 1e-28 | -6.483e+01 | 0.0000 | 3446.0 | 24.06% | 2515.4 | 20.25% | motif file (matrix) |
pdf |
| 24 |  | Sox6(HMG)/Myotubes-Sox6-ChIP-Seq(GSE32627)/Homer | 1e-27 | -6.362e+01 | 0.0000 | 2375.0 | 16.58% | 1659.0 | 13.36% | motif file (matrix) |
pdf |
| 25 |  | Pitx1(Homeobox)/Chicken-Pitx1-ChIP-Seq(GSE38910)/Homer | 1e-26 | -6.145e+01 | 0.0000 | 5812.0 | 40.57% | 4497.5 | 36.21% | motif file (matrix) |
pdf |
| 26 |  | Oct4(POU/Homeobox)/mES-Oct4-ChIP-Seq(GSE11431)/Homer | 1e-26 | -6.122e+01 | 0.0000 | 672.0 | 4.69% | 376.2 | 3.03% | motif file (matrix) |
pdf |
| 27 |  | TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer | 1e-26 | -6.089e+01 | 0.0000 | 935.0 | 6.53% | 564.8 | 4.55% | motif file (matrix) |
pdf |
| 28 |  | Tcf3(HMG)/mES-Tcf3-ChIP-Seq(GSE11724)/Homer | 1e-26 | -6.004e+01 | 0.0000 | 438.0 | 3.06% | 219.3 | 1.77% | motif file (matrix) |
pdf |
| 29 |  | Tcf12(HLH)/GM12878-Tcf12-ChIP-Seq(GSE32465)/Homer | 1e-23 | -5.467e+01 | 0.0000 | 1737.0 | 12.13% | 1185.2 | 9.54% | motif file (matrix) |
pdf |
| 30 |  | Tcf4(HMG)/Hct116-Tcf4-ChIP-Seq(SRA012054)/Homer | 1e-22 | -5.088e+01 | 0.0000 | 738.0 | 5.15% | 440.9 | 3.55% | motif file (matrix) |
pdf |
| 31 |  | Oct2(POU/Homeobox)/Bcell-Oct2-ChIP-Seq(GSE21512)/Homer | 1e-20 | -4.755e+01 | 0.0000 | 403.0 | 2.81% | 211.6 | 1.70% | motif file (matrix) |
pdf |
| 32 |  | Tbx5(T-box)/HL1-Tbx5.biotin-ChIP-Seq(GSE21529)/Homer | 1e-18 | -4.246e+01 | 0.0000 | 5094.0 | 35.56% | 3983.6 | 32.07% | motif file (matrix) |
pdf |
| 33 |  | Phox2a(Homeobox)/Neuron-Phox2a-ChIP-Seq(GSE31456)/Homer | 1e-18 | -4.205e+01 | 0.0000 | 509.0 | 3.55% | 292.5 | 2.35% | motif file (matrix) |
pdf |
| 34 |  | TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer | 1e-16 | -3.827e+01 | 0.0000 | 1195.0 | 8.34% | 812.0 | 6.54% | motif file (matrix) |
pdf |
| 35 |  | Myf5(bHLH)/GM-Myf5-ChIP-Seq(GSE24852)/Homer | 1e-15 | -3.681e+01 | 0.0000 | 1223.0 | 8.54% | 838.9 | 6.75% | motif file (matrix) |
pdf |
| 36 |  | E2A(HLH)/proBcell-E2A-ChIP-Seq(GSE21978)/Homer | 1e-14 | -3.354e+01 | 0.0000 | 2447.0 | 17.08% | 1828.1 | 14.72% | motif file (matrix) |
pdf |
| 37 | 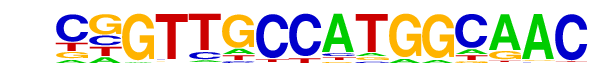 | RFX(HTH)/K562-RFX3-ChIP-Seq(SRA012198)/Homer | 1e-13 | -3.206e+01 | 0.0000 | 189.0 | 1.32% | 89.9 | 0.72% | motif file (matrix) |
pdf |
| 38 |  | NFY(CCAAT)/Promoter/Homer | 1e-13 | -3.144e+01 | 0.0000 | 997.0 | 6.96% | 679.4 | 5.47% | motif file (matrix) |
pdf |
| 39 |  | Ap4(HLH)/AML-Tfap4-ChIP-Seq(GSE45738)/Homer | 1e-13 | -3.038e+01 | 0.0000 | 2047.0 | 14.29% | 1517.2 | 12.21% | motif file (matrix) |
pdf |
| 40 |  | HNF6(Homeobox)/Liver-Hnf6-ChIP-Seq(ERP000394)/Homer | 1e-12 | -2.926e+01 | 0.0000 | 667.0 | 4.66% | 433.7 | 3.49% | motif file (matrix) |
pdf |
| 41 |  | Rfx1(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-11 | -2.621e+01 | 0.0000 | 344.0 | 2.40% | 202.1 | 1.63% | motif file (matrix) |
pdf |
| 42 |  | Sp1(Zf)/Promoter/Homer | 1e-10 | -2.473e+01 | 0.0000 | 434.0 | 3.03% | 270.9 | 2.18% | motif file (matrix) |
pdf |
| 43 |  | Hnf1(Homeobox)/Liver-Foxa2-Chip-Seq(GSE25694)/Homer | 1e-10 | -2.412e+01 | 0.0000 | 154.0 | 1.08% | 75.7 | 0.61% | motif file (matrix) |
pdf |
| 44 |  | OCT4-SOX2-TCF-NANOG((POU/Homeobox/HMG)/mES-Oct4-ChIP-Seq(GSE11431)/Homer | 1e-10 | -2.396e+01 | 0.0000 | 220.0 | 1.54% | 119.8 | 0.96% | motif file (matrix) |
pdf |
| 45 |  | TCFL2(HMG)/K562-TCF7L2-ChIP-Seq(GSE29196)/Homer | 1e-9 | -2.256e+01 | 0.0000 | 159.0 | 1.11% | 80.7 | 0.65% | motif file (matrix) |
pdf |
| 46 |  | Rfx2(HTH)/LoVo-RFX2-ChIP-Seq(GSE49402)/Homer | 1e-9 | -2.237e+01 | 0.0000 | 205.0 | 1.43% | 111.6 | 0.90% | motif file (matrix) |
pdf |
| 47 |  | Rfx5(HTH)/GM12878-Rfx5-ChIP-Seq(GSE31477)/Homer | 1e-9 | -2.171e+01 | 0.0000 | 537.0 | 3.75% | 354.1 | 2.85% | motif file (matrix) |
pdf |
| 48 |  | CTCF-SatelliteElement/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-9 | -2.126e+01 | 0.0000 | 38.0 | 0.27% | 10.2 | 0.08% | motif file (matrix) |
pdf |
| 49 |  | Nur77(NR)/K562-NR4A1-ChIP-Seq(GSE31363)/Homer | 1e-8 | -2.055e+01 | 0.0000 | 278.0 | 1.94% | 165.5 | 1.33% | motif file (matrix) |
pdf |
| 50 |  | MyoD(HLH)/Myotube-MyoD-ChIP-Seq(GSE21614)/Homer | 1e-8 | -2.049e+01 | 0.0000 | 1288.0 | 8.99% | 948.4 | 7.64% | motif file (matrix) |
pdf |
| 51 |  | GATA:SCL/Ter119-SCL-ChIP-Seq(GSE18720)/Homer | 1e-8 | -1.980e+01 | 0.0000 | 201.0 | 1.40% | 112.2 | 0.90% | motif file (matrix) |
pdf |
| 52 |  | X-box(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-8 | -1.944e+01 | 0.0000 | 216.0 | 1.51% | 123.6 | 1.00% | motif file (matrix) |
pdf |
| 53 |  | Gfi1b(Zf)/HPC7-Gfi1b-ChIP-Seq(GSE22178)/Homer | 1e-8 | -1.929e+01 | 0.0000 | 859.0 | 6.00% | 611.0 | 4.92% | motif file (matrix) |
pdf |
| 54 |  | KLF5(Zf)/LoVo-KLF5-ChIP-Seq(GSE49402)/Homer | 1e-8 | -1.875e+01 | 0.0000 | 1942.0 | 13.56% | 1489.5 | 11.99% | motif file (matrix) |
pdf |
| 55 |  | Oct4:Sox17/F9-Sox17-ChIP-Seq(GSE44553)/Homer | 1e-7 | -1.833e+01 | 0.0000 | 212.0 | 1.48% | 122.4 | 0.99% | motif file (matrix) |
pdf |
| 56 |  | EBF1(EBF)/Near-E2A-ChIP-Seq(GSE21512)/Homer | 1e-7 | -1.747e+01 | 0.0000 | 1648.0 | 11.50% | 1255.3 | 10.11% | motif file (matrix) |
pdf |
| 57 |  | Pax7-long(Paired/Homeobox)/Myoblast-Pax7-ChIP-Seq(GSE25064)/Homer | 1e-7 | -1.655e+01 | 0.0000 | 38.0 | 0.27% | 12.9 | 0.10% | motif file (matrix) |
pdf |
| 58 |  | Ptf1a(HLH)/Panc1-Ptf1a-ChIP-Seq(GSE47459)/Homer | 1e-7 | -1.652e+01 | 0.0000 | 4165.0 | 29.08% | 3366.8 | 27.11% | motif file (matrix) |
pdf |
| 59 |  | MyoG(HLH)/C2C12-MyoG-ChIP-Seq(GSE36024)/Homer | 1e-7 | -1.640e+01 | 0.0000 | 1782.0 | 12.44% | 1371.7 | 11.04% | motif file (matrix) |
pdf |
| 60 |  | NeuroD1(bHLH)/Islet-NeuroD1-ChIP-Seq(GSE30298)/Homer | 1e-6 | -1.597e+01 | 0.0000 | 1226.0 | 8.56% | 919.9 | 7.41% | motif file (matrix) |
pdf |
| 61 |  | Tbox:Smad/ESCd5-Smad2_3-ChIP-Seq(GSE29422)/Homer | 1e-6 | -1.562e+01 | 0.0000 | 312.0 | 2.18% | 200.1 | 1.61% | motif file (matrix) |
pdf |
| 62 |  | REST-NRSF(Zf)/Jurkat-NRSF-ChIP-Seq/Homer | 1e-6 | -1.557e+01 | 0.0000 | 31.0 | 0.22% | 9.3 | 0.07% | motif file (matrix) |
pdf |
| 63 |  | Foxh1(Forkhead)/hESC-FOXH1-ChIP-Seq(GSE29422)/Homer | 1e-6 | -1.425e+01 | 0.0000 | 820.0 | 5.72% | 600.2 | 4.83% | motif file (matrix) |
pdf |
| 64 |  | CEBP:CEBP(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer | 1e-5 | -1.271e+01 | 0.0000 | 142.0 | 0.99% | 82.7 | 0.67% | motif file (matrix) |
pdf |
| 65 |  | E-box(HLH)/Promoter/Homer | 1e-5 | -1.221e+01 | 0.0000 | 99.0 | 0.69% | 53.1 | 0.43% | motif file (matrix) |
pdf |
| 66 |  | GATA-IR4(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-5 | -1.208e+01 | 0.0000 | 109.0 | 0.76% | 60.6 | 0.49% | motif file (matrix) |
pdf |
| 67 |  | Pax7(Paired/Homeobox)/Myoblast-Pax7-ChIP-Seq(GSE25064)/Homer | 1e-5 | -1.166e+01 | 0.0000 | 111.0 | 0.77% | 62.9 | 0.51% | motif file (matrix) |
pdf |
| 68 |  | EBF(EBF)/proBcell-EBF-ChIP-Seq(GSE21978)/Homer | 1e-4 | -1.147e+01 | 0.0000 | 328.0 | 2.29% | 223.4 | 1.80% | motif file (matrix) |
pdf |
| 69 |  | PU.1(ETS)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-4 | -1.125e+01 | 0.0000 | 706.0 | 4.93% | 522.4 | 4.21% | motif file (matrix) |
pdf |
| 70 |  | E2A-nearPU.1(HLH)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-4 | -1.095e+01 | 0.0001 | 2187.0 | 15.27% | 1745.5 | 14.05% | motif file (matrix) |
pdf |
| 71 |  | Hoxb4(Homeobox)/ES-Hoxb4-ChIP-Seq(GSE34014)/Homer | 1e-4 | -9.777e+00 | 0.0002 | 238.0 | 1.66% | 160.0 | 1.29% | motif file (matrix) |
pdf |
| 72 |  | PAX5-shortForm(Paired/Homeobox)/GM12878-PAX5-ChIP-Seq(GSE32465)/Homer | 1e-4 | -9.614e+00 | 0.0002 | 119.0 | 0.83% | 71.1 | 0.57% | motif file (matrix) |
pdf |
| 73 |  | Srebp2(HLH)/HepG2-Srebp2-ChIP-Seq(GSE31477)/Homer | 1e-4 | -9.582e+00 | 0.0002 | 187.0 | 1.31% | 121.0 | 0.97% | motif file (matrix) |
pdf |
| 74 |  | FOXA1:AR/LNCAP-AR-ChIP-Seq(GSE27824)/Homer | 1e-4 | -9.496e+00 | 0.0002 | 89.0 | 0.62% | 50.1 | 0.40% | motif file (matrix) |
pdf |
| 75 |  | ZNF143|STAF(Zf)/CUTLL-ZNF143-ChIP-Seq(GSE29600)/Homer | 1e-3 | -9.110e+00 | 0.0004 | 478.0 | 3.34% | 349.9 | 2.82% | motif file (matrix) |
pdf |
| 76 |  | TATA-Box(TBP)/Promoter/Homer | 1e-3 | -8.612e+00 | 0.0006 | 1584.0 | 11.06% | 1260.3 | 10.15% | motif file (matrix) |
pdf |
| 77 |  | GFX(?)/Promoter/Homer | 1e-3 | -8.528e+00 | 0.0006 | 7.0 | 0.05% | 1.9 | 0.02% | motif file (matrix) |
pdf |
| 78 |  | CHR/Cell-Cycle-Exp/Homer | 1e-3 | -8.316e+00 | 0.0008 | 683.0 | 4.77% | 518.0 | 4.17% | motif file (matrix) |
pdf |
| 79 |  | NFkB-p65(RHD)/GM12787-p65-ChIP-Seq(GSE19485)/Homer | 1e-3 | -8.141e+00 | 0.0009 | 656.0 | 4.58% | 497.2 | 4.00% | motif file (matrix) |
pdf |
| 80 |  | Pdx1(Homeobox)/Islet-Pdx1-ChIP-Seq(SRA008281)/Homer | 1e-3 | -7.763e+00 | 0.0013 | 1077.0 | 7.52% | 845.6 | 6.81% | motif file (matrix) |
pdf |
| 81 |  | GATA-DR8(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-3 | -7.190e+00 | 0.0023 | 71.0 | 0.50% | 41.3 | 0.33% | motif file (matrix) |
pdf |
| 82 |  | Six1(Homeobox)/Myoblast-Six1-ChIP-Chip(GSE20150)/Homer | 1e-3 | -7.050e+00 | 0.0026 | 281.0 | 1.96% | 201.6 | 1.62% | motif file (matrix) |
pdf |
| 83 |  | PBX1(Homeobox)/MCF7-PBX1-ChIP-Seq(GSE28007)/Homer | 1e-2 | -6.766e+00 | 0.0034 | 155.0 | 1.08% | 104.2 | 0.84% | motif file (matrix) |
pdf |
| 84 |  | MafK(bZIP)/C2C12-MafK-ChIP-Seq(GSE36030)/Homer | 1e-2 | -6.661e+00 | 0.0037 | 327.0 | 2.28% | 239.3 | 1.93% | motif file (matrix) |
pdf |
| 85 |  | Tbx20(T-box)/Heart-Tbx20-ChIP-Seq(GSE29636)/Homer | 1e-2 | -6.561e+00 | 0.0041 | 349.0 | 2.44% | 257.4 | 2.07% | motif file (matrix) |
pdf |
| 86 |  | TR4(NR/DR1)/Hela-TR4-ChIP-Seq(GSE24685)/Homer | 1e-2 | -6.300e+00 | 0.0053 | 152.0 | 1.06% | 103.2 | 0.83% | motif file (matrix) |
pdf |
| 87 |  | Srebp1a(HLH)/HepG2-Srebp1a-ChIP-Seq(GSE31477)/Homer | 1e-2 | -6.142e+00 | 0.0061 | 269.0 | 1.88% | 195.7 | 1.58% | motif file (matrix) |
pdf |
| 88 |  | Atoh1(bHLH)/Cerebellum-Atoh1-ChIP-Seq(GSE22111)/Homer | 1e-2 | -5.691e+00 | 0.0094 | 1639.0 | 11.44% | 1333.4 | 10.74% | motif file (matrix) |
pdf |
| 89 |  | Maz(Zf)/HepG2-Maz-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.053e+00 | 0.0177 | 2136.0 | 14.91% | 1761.2 | 14.18% | motif file (matrix) |
pdf |
| 90 |  | PAX3:FKHR-fusion(Paired/Homeobox)/Rh4-PAX3:FKHR-ChIP-Seq(GSE19063)/Homer | 1e-2 | -5.052e+00 | 0.0177 | 232.0 | 1.62% | 171.0 | 1.38% | motif file (matrix) |
pdf |
| 91 |  | CArG(MADS)/PUER-Srf-ChIP-Seq(Sullivan et al.)/Homer | 1e-2 | -4.960e+00 | 0.0190 | 342.0 | 2.39% | 259.7 | 2.09% | motif file (matrix) |
pdf |
| 92 |  | ETS:E-box/HPC7-Scl-ChIP-Seq(GSE22178)/Homer | 1e-2 | -4.878e+00 | 0.0204 | 131.0 | 0.91% | 91.7 | 0.74% | motif file (matrix) |
pdf |
| 93 |  | Unknown(Homeobox)/Limb-p300-ChIP-Seq/Homer | 1e-2 | -4.740e+00 | 0.0231 | 688.0 | 4.80% | 545.6 | 4.39% | motif file (matrix) |
pdf |

























































































































































































