| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-197 | -4.550e+02 | 0.0000 | 1685.0 | 17.59% | 593.5 | 8.07% | motif file (matrix) |
pdf |
| 2 |  | FOXA1(Forkhead)/MCF7-FOXA1-ChIP-Seq(GSE26831)/Homer | 1e-182 | -4.204e+02 | 0.0000 | 1415.0 | 14.77% | 472.3 | 6.42% | motif file (matrix) |
pdf |
| 3 |  | Foxa2(Forkhead)/Liver-Foxa2-ChIP-Seq(GSE25694)/Homer | 1e-141 | -3.258e+02 | 0.0000 | 1211.0 | 12.64% | 421.9 | 5.73% | motif file (matrix) |
pdf |
| 4 |  | Fox:Ebox(Forkhead:HLH)/Panc1-Foxa2-ChIP-Seq(GSE47459)/Homer | 1e-122 | -2.831e+02 | 0.0000 | 1496.0 | 15.62% | 606.9 | 8.25% | motif file (matrix) |
pdf |
| 5 |  | CTCF(Zf)/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-86 | -1.994e+02 | 0.0000 | 394.0 | 4.11% | 93.3 | 1.27% | motif file (matrix) |
pdf |
| 6 | 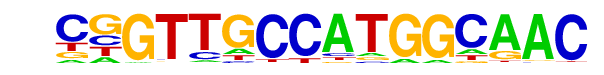 | RFX(HTH)/K562-RFX3-ChIP-Seq(SRA012198)/Homer | 1e-75 | -1.737e+02 | 0.0000 | 167.0 | 1.74% | 20.6 | 0.28% | motif file (matrix) |
pdf |
| 7 |  | BORIS(Zf)/K562-CTCFL-ChIP-Seq(GSE32465)/Homer | 1e-74 | -1.707e+02 | 0.0000 | 484.0 | 5.05% | 145.0 | 1.97% | motif file (matrix) |
pdf |
| 8 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-63 | -1.451e+02 | 0.0000 | 581.0 | 6.06% | 206.2 | 2.80% | motif file (matrix) |
pdf |
| 9 |  | Rfx2(HTH)/LoVo-RFX2-ChIP-Seq(GSE49402)/Homer | 1e-61 | -1.416e+02 | 0.0000 | 179.0 | 1.87% | 29.8 | 0.41% | motif file (matrix) |
pdf |
| 10 |  | TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer | 1e-52 | -1.218e+02 | 0.0000 | 981.0 | 10.24% | 451.4 | 6.13% | motif file (matrix) |
pdf |
| 11 |  | Sox3(HMG)/NPC-Sox3-ChIP-Seq(GSE33059)/Homer | 1e-48 | -1.109e+02 | 0.0000 | 1978.0 | 20.65% | 1107.4 | 15.05% | motif file (matrix) |
pdf |
| 12 |  | Gata4(Zf)/Heart-Gata4-ChIP-Seq(GSE35151)/Homer | 1e-47 | -1.105e+02 | 0.0000 | 1003.0 | 10.47% | 477.9 | 6.50% | motif file (matrix) |
pdf |
| 13 |  | TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer | 1e-47 | -1.104e+02 | 0.0000 | 734.0 | 7.66% | 317.1 | 4.31% | motif file (matrix) |
pdf |
| 14 |  | Sox2(HMG)/mES-Sox2-ChIP-Seq(GSE11431)/Homer | 1e-43 | -1.008e+02 | 0.0000 | 1084.0 | 11.32% | 539.6 | 7.33% | motif file (matrix) |
pdf |
| 15 |  | Foxo1(Forkhead)/RAW-Foxo1-ChIP-Seq(Fan et al.)/Homer | 1e-41 | -9.564e+01 | 0.0000 | 2085.0 | 21.76% | 1208.2 | 16.42% | motif file (matrix) |
pdf |
| 16 |  | Gata2(Zf)/K562-GATA2-ChIP-Seq(GSE18829)/Homer | 1e-41 | -9.554e+01 | 0.0000 | 704.0 | 7.35% | 314.6 | 4.28% | motif file (matrix) |
pdf |
| 17 |  | Gata1(Zf)/K562-GATA1-ChIP-Seq(GSE18829)/Homer | 1e-36 | -8.388e+01 | 0.0000 | 636.0 | 6.64% | 286.2 | 3.89% | motif file (matrix) |
pdf |
| 18 |  | GATA3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-34 | -7.869e+01 | 0.0000 | 1388.0 | 14.49% | 769.2 | 10.46% | motif file (matrix) |
pdf |
| 19 |  | Sox6(HMG)/Myotubes-Sox6-ChIP-Seq(GSE32627)/Homer | 1e-30 | -7.016e+01 | 0.0000 | 1694.0 | 17.68% | 992.2 | 13.49% | motif file (matrix) |
pdf |
| 20 |  | X-box(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-30 | -6.963e+01 | 0.0000 | 180.0 | 1.88% | 51.7 | 0.70% | motif file (matrix) |
pdf |
| 21 |  | Rfx5(HTH)/GM12878-Rfx5-ChIP-Seq(GSE31477)/Homer | 1e-22 | -5.189e+01 | 0.0000 | 390.0 | 4.07% | 175.6 | 2.39% | motif file (matrix) |
pdf |
| 22 |  | Rfx1(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-21 | -5.008e+01 | 0.0000 | 271.0 | 2.83% | 109.1 | 1.48% | motif file (matrix) |
pdf |
| 23 |  | Lhx3(Homeobox)/Neuron-Lhx3-ChIP-Seq(GSE31456)/Homer | 1e-20 | -4.769e+01 | 0.0000 | 1396.0 | 14.57% | 838.9 | 11.40% | motif file (matrix) |
pdf |
| 24 |  | GRHL2(CP2)/HBE-GRHL2-ChIP-Seq(GSE46194)/Homer | 1e-19 | -4.530e+01 | 0.0000 | 361.0 | 3.77% | 165.2 | 2.24% | motif file (matrix) |
pdf |
| 25 |  | Eomes(T-box)/H9-Eomes-ChIP-Seq(GSE26097)/Homer | 1e-17 | -4.106e+01 | 0.0000 | 1934.0 | 20.19% | 1234.6 | 16.78% | motif file (matrix) |
pdf |
| 26 |  | Nkx6.1(Homeobox)/Islet-Nkx6.1-ChIP-Seq(GSE40975)/Homer | 1e-17 | -4.030e+01 | 0.0000 | 2016.0 | 21.04% | 1295.2 | 17.60% | motif file (matrix) |
pdf |
| 27 |  | Oct2(POU/Homeobox)/Bcell-Oct2-ChIP-Seq(GSE21512)/Homer | 1e-16 | -3.866e+01 | 0.0000 | 221.0 | 2.31% | 91.7 | 1.25% | motif file (matrix) |
pdf |
| 28 |  | Tcf4(HMG)/Hct116-Tcf4-ChIP-Seq(SRA012054)/Homer | 1e-16 | -3.826e+01 | 0.0000 | 501.0 | 5.23% | 260.7 | 3.54% | motif file (matrix) |
pdf |
| 29 |  | CRX(Homeobox)/Retina-Crx-ChIP-Seq(GSE20012)/Homer | 1e-16 | -3.757e+01 | 0.0000 | 2005.0 | 20.93% | 1296.2 | 17.62% | motif file (matrix) |
pdf |
| 30 |  | Sp1(Zf)/Promoter/Homer | 1e-15 | -3.619e+01 | 0.0000 | 402.0 | 4.20% | 201.1 | 2.73% | motif file (matrix) |
pdf |
| 31 |  | Pitx1(Homeobox)/Chicken-Pitx1-ChIP-Seq(GSE38910)/Homer | 1e-15 | -3.609e+01 | 0.0000 | 3580.0 | 37.37% | 2458.3 | 33.41% | motif file (matrix) |
pdf |
| 32 |  | Lhx2(Homeobox)/HFSC-Lhx2-ChIP-Seq(GSE48068)/Homer | 1e-15 | -3.584e+01 | 0.0000 | 909.0 | 9.49% | 533.6 | 7.25% | motif file (matrix) |
pdf |
| 33 |  | Oct4(POU/Homeobox)/mES-Oct4-ChIP-Seq(GSE11431)/Homer | 1e-13 | -3.089e+01 | 0.0000 | 345.0 | 3.60% | 173.4 | 2.36% | motif file (matrix) |
pdf |
| 34 |  | Unknown-ESC-element/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-12 | -2.772e+01 | 0.0000 | 766.0 | 8.00% | 455.3 | 6.19% | motif file (matrix) |
pdf |
| 35 |  | Tcf3(HMG)/mES-Tcf3-ChIP-Seq(GSE11724)/Homer | 1e-11 | -2.625e+01 | 0.0000 | 291.0 | 3.04% | 146.9 | 2.00% | motif file (matrix) |
pdf |
| 36 |  | TCFL2(HMG)/K562-TCF7L2-ChIP-Seq(GSE29196)/Homer | 1e-11 | -2.577e+01 | 0.0000 | 112.0 | 1.17% | 42.8 | 0.58% | motif file (matrix) |
pdf |
| 37 |  | Nanog(Homeobox)/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-9 | -2.299e+01 | 0.0000 | 3619.0 | 37.78% | 2550.1 | 34.66% | motif file (matrix) |
pdf |
| 38 |  | Six1(Homeobox)/Myoblast-Six1-ChIP-Chip(GSE20150)/Homer | 1e-9 | -2.276e+01 | 0.0000 | 233.0 | 2.43% | 115.0 | 1.56% | motif file (matrix) |
pdf |
| 39 |  | SPDEF(ETS)/VCaP-SPDEF-ChIP-Seq(SRA014231)/Homer | 1e-9 | -2.139e+01 | 0.0000 | 1027.0 | 10.72% | 654.8 | 8.90% | motif file (matrix) |
pdf |
| 40 |  | Tbx5(T-box)/HL1-Tbx5.biotin-ChIP-Seq(GSE21529)/Homer | 1e-8 | -2.001e+01 | 0.0000 | 3143.0 | 32.81% | 2209.7 | 30.03% | motif file (matrix) |
pdf |
| 41 |  | NRF1(NRF)/MCF7-NRF1-ChIP-Seq(Unpublished)/Homer | 1e-8 | -1.997e+01 | 0.0000 | 139.0 | 1.45% | 62.2 | 0.85% | motif file (matrix) |
pdf |
| 42 |  | KLF5(Zf)/LoVo-KLF5-ChIP-Seq(GSE49402)/Homer | 1e-8 | -1.955e+01 | 0.0000 | 1562.0 | 16.30% | 1044.7 | 14.20% | motif file (matrix) |
pdf |
| 43 |  | Foxh1(Forkhead)/hESC-FOXH1-ChIP-Seq(GSE29422)/Homer | 1e-7 | -1.812e+01 | 0.0000 | 487.0 | 5.08% | 289.1 | 3.93% | motif file (matrix) |
pdf |
| 44 |  | PRDM1/BMI1(Zf)/Hela-PRDM1-ChIP-Seq(GSE31477)/Homer | 1e-7 | -1.644e+01 | 0.0000 | 444.0 | 4.63% | 264.8 | 3.60% | motif file (matrix) |
pdf |
| 45 |  | Hnf1(Homeobox)/Liver-Foxa2-Chip-Seq(GSE25694)/Homer | 1e-6 | -1.595e+01 | 0.0000 | 118.0 | 1.23% | 54.8 | 0.75% | motif file (matrix) |
pdf |
| 46 |  | c-Jun-CRE(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-6 | -1.576e+01 | 0.0000 | 243.0 | 2.54% | 132.5 | 1.80% | motif file (matrix) |
pdf |
| 47 |  | OCT4-SOX2-TCF-NANOG((POU/Homeobox/HMG)/mES-Oct4-ChIP-Seq(GSE11431)/Homer | 1e-6 | -1.570e+01 | 0.0000 | 109.0 | 1.14% | 49.4 | 0.67% | motif file (matrix) |
pdf |
| 48 |  | NRF1/Promoter/Homer | 1e-6 | -1.533e+01 | 0.0000 | 163.0 | 1.70% | 82.8 | 1.13% | motif file (matrix) |
pdf |
| 49 |  | Phox2a(Homeobox)/Neuron-Phox2a-ChIP-Seq(GSE31456)/Homer | 1e-6 | -1.463e+01 | 0.0000 | 246.0 | 2.57% | 136.8 | 1.86% | motif file (matrix) |
pdf |
| 50 |  | Gfi1b(Zf)/HPC7-Gfi1b-ChIP-Seq(GSE22178)/Homer | 1e-6 | -1.419e+01 | 0.0000 | 527.0 | 5.50% | 327.5 | 4.45% | motif file (matrix) |
pdf |
| 51 |  | HNF6(Homeobox)/Liver-Hnf6-ChIP-Seq(ERP000394)/Homer | 1e-5 | -1.373e+01 | 0.0000 | 347.0 | 3.62% | 206.0 | 2.80% | motif file (matrix) |
pdf |
| 52 | 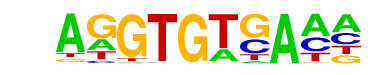 | Tbet(T-box)/CD8-Tbet-ChIP-Seq(GSE33802)/Homer | 1e-5 | -1.324e+01 | 0.0000 | 952.0 | 9.94% | 631.5 | 8.58% | motif file (matrix) |
pdf |
| 53 |  | JunD(bZIP)/K562-JunD-ChIP-Seq/Homer | 1e-5 | -1.300e+01 | 0.0000 | 87.0 | 0.91% | 39.3 | 0.53% | motif file (matrix) |
pdf |
| 54 |  | Oct4:Sox17/F9-Sox17-ChIP-Seq(GSE44553)/Homer | 1e-5 | -1.287e+01 | 0.0000 | 130.0 | 1.36% | 65.3 | 0.89% | motif file (matrix) |
pdf |
| 55 |  | CArG(MADS)/PUER-Srf-ChIP-Seq(Sullivan et al.)/Homer | 1e-5 | -1.285e+01 | 0.0000 | 227.0 | 2.37% | 127.3 | 1.73% | motif file (matrix) |
pdf |
| 56 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-5 | -1.247e+01 | 0.0000 | 595.0 | 6.21% | 380.9 | 5.18% | motif file (matrix) |
pdf |
| 57 |  | NF1:FOXA1/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-5 | -1.190e+01 | 0.0000 | 57.0 | 0.59% | 23.5 | 0.32% | motif file (matrix) |
pdf |
| 58 |  | ETS(ETS)/Promoter/Homer | 1e-5 | -1.168e+01 | 0.0000 | 357.0 | 3.73% | 217.5 | 2.96% | motif file (matrix) |
pdf |
| 59 |  | Egr2/Thymocytes-Egr2-ChIP-Seq(GSE34254)/Homer | 1e-4 | -1.139e+01 | 0.0000 | 186.0 | 1.94% | 103.3 | 1.40% | motif file (matrix) |
pdf |
| 60 |  | Znf263(Zf)/K562-Znf263-ChIP-Seq(GSE31477)/Homer | 1e-4 | -1.078e+01 | 0.0001 | 2141.0 | 22.35% | 1518.4 | 20.64% | motif file (matrix) |
pdf |
| 61 |  | Pdx1(Homeobox)/Islet-Pdx1-ChIP-Seq(SRA008281)/Homer | 1e-4 | -1.078e+01 | 0.0001 | 616.0 | 6.43% | 401.2 | 5.45% | motif file (matrix) |
pdf |
| 62 |  | GFY(?)/Promoter/Homer | 1e-4 | -1.075e+01 | 0.0001 | 48.0 | 0.50% | 19.0 | 0.26% | motif file (matrix) |
pdf |
| 63 |  | Hoxb4(Homeobox)/ES-Hoxb4-ChIP-Seq(GSE34014)/Homer | 1e-4 | -1.071e+01 | 0.0001 | 125.0 | 1.30% | 65.2 | 0.89% | motif file (matrix) |
pdf |
| 64 |  | CTCF-SatelliteElement/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-4 | -1.029e+01 | 0.0001 | 26.0 | 0.27% | 8.7 | 0.12% | motif file (matrix) |
pdf |
| 65 |  | Stat3(Stat)/mES-Stat3-ChIP-Seq(GSE11431)/Homer | 1e-4 | -9.747e+00 | 0.0002 | 455.0 | 4.75% | 291.7 | 3.96% | motif file (matrix) |
pdf |
| 66 |  | Pax7-long(Paired/Homeobox)/Myoblast-Pax7-ChIP-Seq(GSE25064)/Homer | 1e-4 | -9.439e+00 | 0.0003 | 23.0 | 0.24% | 7.6 | 0.10% | motif file (matrix) |
pdf |
| 67 |  | REST-NRSF(Zf)/Jurkat-NRSF-ChIP-Seq/Homer | 1e-4 | -9.439e+00 | 0.0003 | 23.0 | 0.24% | 7.9 | 0.11% | motif file (matrix) |
pdf |
| 68 |  | GFY-Staf/Promoters/Homer | 1e-3 | -8.611e+00 | 0.0007 | 32.0 | 0.33% | 12.3 | 0.17% | motif file (matrix) |
pdf |
| 69 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-3 | -8.465e+00 | 0.0008 | 565.0 | 5.90% | 374.8 | 5.09% | motif file (matrix) |
pdf |
| 70 |  | Isl1(Homeobox)/Neuron-Isl1-ChIP-Seq(GSE31456)/Homer | 1e-3 | -8.463e+00 | 0.0008 | 1669.0 | 17.42% | 1183.0 | 16.08% | motif file (matrix) |
pdf |
| 71 | 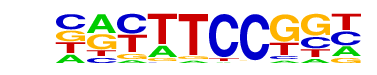 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-3 | -8.318e+00 | 0.0008 | 1154.0 | 12.05% | 803.8 | 10.93% | motif file (matrix) |
pdf |
| 72 |  | Klf4(Zf)/mES-Klf4-ChIP-Seq(GSE11431)/Homer | 1e-3 | -8.267e+00 | 0.0009 | 453.0 | 4.73% | 295.2 | 4.01% | motif file (matrix) |
pdf |
| 73 |  | EBF1(EBF)/Near-E2A-ChIP-Seq(GSE21512)/Homer | 1e-3 | -8.139e+00 | 0.0010 | 1162.0 | 12.13% | 810.2 | 11.01% | motif file (matrix) |
pdf |
| 74 |  | E2A(HLH)/proBcell-E2A-ChIP-Seq(GSE21978)/Homer | 1e-3 | -7.861e+00 | 0.0013 | 1616.0 | 16.87% | 1148.2 | 15.61% | motif file (matrix) |
pdf |
| 75 |  | IRF2(IRF)/Erythroblas-IRF2-ChIP-Seq(GSE36985)/Homer | 1e-3 | -7.680e+00 | 0.0015 | 78.0 | 0.81% | 40.6 | 0.55% | motif file (matrix) |
pdf |
| 76 |  | GATA-IR4(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-3 | -7.435e+00 | 0.0019 | 58.0 | 0.61% | 28.7 | 0.39% | motif file (matrix) |
pdf |
| 77 |  | Srebp1a(HLH)/HepG2-Srebp1a-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.173e+00 | 0.0025 | 171.0 | 1.78% | 102.3 | 1.39% | motif file (matrix) |
pdf |
| 78 |  | STAT1(Stat)/HelaS3-STAT1-ChIP-Seq(GSE12782)/Homer | 1e-2 | -6.697e+00 | 0.0039 | 226.0 | 2.36% | 141.1 | 1.92% | motif file (matrix) |
pdf |
| 79 |  | RUNX(Runt)/HPC7-Runx1-ChIP-Seq(GSE22178)/Homer | 1e-2 | -6.663e+00 | 0.0040 | 635.0 | 6.63% | 433.3 | 5.89% | motif file (matrix) |
pdf |
| 80 |  | RUNX-AML(Runt)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer | 1e-2 | -6.627e+00 | 0.0041 | 643.0 | 6.71% | 439.5 | 5.97% | motif file (matrix) |
pdf |
| 81 |  | SCL(HLH)/HPC7-Scl-ChIP-Seq(GSE13511)/Homer | 1e-2 | -6.486e+00 | 0.0046 | 4626.0 | 48.29% | 3441.7 | 46.78% | motif file (matrix) |
pdf |
| 82 | 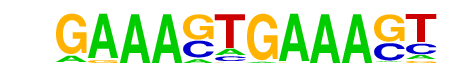 | IRF1(IRF)/PBMC-IRF1-ChIP-Seq(GSE43036)/Homer | 1e-2 | -6.329e+00 | 0.0054 | 110.0 | 1.15% | 63.2 | 0.86% | motif file (matrix) |
pdf |
| 83 |  | Maz(Zf)/HepG2-Maz-ChIP-Seq(GSE31477)/Homer | 1e-2 | -6.142e+00 | 0.0064 | 1818.0 | 18.98% | 1313.9 | 17.86% | motif file (matrix) |
pdf |
| 84 |  | Arnt:Ahr(bHLH)/MCF7-Arnt-ChIP-Seq(Lo et al.)/Homer | 1e-2 | -6.103e+00 | 0.0066 | 511.0 | 5.33% | 346.8 | 4.71% | motif file (matrix) |
pdf |
| 85 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-2 | -6.006e+00 | 0.0071 | 560.0 | 5.85% | 382.4 | 5.20% | motif file (matrix) |
pdf |
| 86 |  | bZIP:IRF/Th17-BatF-ChIP-Seq(GSE39756)/Homer | 1e-2 | -5.946e+00 | 0.0075 | 281.0 | 2.93% | 182.2 | 2.48% | motif file (matrix) |
pdf |
| 87 |  | LXRE(NR/DR4)/BLRP(RAW)-LXRb-ChIP-Seq(GSE21512)/Homer | 1e-2 | -5.934e+00 | 0.0075 | 50.0 | 0.52% | 25.2 | 0.34% | motif file (matrix) |
pdf |
| 88 |  | ERE(NR/IR3)/MCF7-ERa-ChIP-Seq(Unpublished)/Homer | 1e-2 | -5.931e+00 | 0.0075 | 224.0 | 2.34% | 142.5 | 1.94% | motif file (matrix) |
pdf |
| 89 |  | PAX3:FKHR-fusion(Paired/Homeobox)/Rh4-PAX3:FKHR-ChIP-Seq(GSE19063)/Homer | 1e-2 | -5.921e+00 | 0.0075 | 134.0 | 1.40% | 81.0 | 1.10% | motif file (matrix) |
pdf |
| 90 |  | GATA-IR3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-2 | -5.873e+00 | 0.0077 | 95.0 | 0.99% | 54.4 | 0.74% | motif file (matrix) |
pdf |
| 91 |  | EBF(EBF)/proBcell-EBF-ChIP-Seq(GSE21978)/Homer | 1e-2 | -5.633e+00 | 0.0097 | 234.0 | 2.44% | 150.1 | 2.04% | motif file (matrix) |
pdf |
| 92 |  | MYB(HTH)/ERMYB-Myb-ChIPSeq(GSE22095)/Homer | 1e-2 | -5.627e+00 | 0.0097 | 1506.0 | 15.72% | 1084.7 | 14.74% | motif file (matrix) |
pdf |
| 93 |  | PPARE(NR/DR1)/3T3L1-Pparg-ChIP-Seq(GSE13511)/Homer | 1e-2 | -5.600e+00 | 0.0098 | 891.0 | 9.30% | 627.7 | 8.53% | motif file (matrix) |
pdf |
| 94 |  | E2F4(E2F)/K562-E2F4-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.498e+00 | 0.0107 | 446.0 | 4.66% | 302.4 | 4.11% | motif file (matrix) |
pdf |
| 95 |  | MafA(bZIP)/Islet-MafA-ChIP-Seq(GSE30298)/Homer | 1e-2 | -5.444e+00 | 0.0112 | 762.0 | 7.95% | 533.9 | 7.26% | motif file (matrix) |
pdf |
| 96 |  | Nkx2.1(Homeobox)/LungAC-Nkx2.1-ChIP-Seq(GSE43252)/Homer | 1e-2 | -5.417e+00 | 0.0114 | 2640.0 | 27.56% | 1940.9 | 26.38% | motif file (matrix) |
pdf |
| 97 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-2 | -5.355e+00 | 0.0120 | 474.0 | 4.95% | 323.8 | 4.40% | motif file (matrix) |
pdf |
| 98 |  | Stat3+il21(Stat)/CD4-Stat3-ChIP-Seq(GSE19198)/Homer | 1e-2 | -5.346e+00 | 0.0120 | 603.0 | 6.29% | 417.9 | 5.68% | motif file (matrix) |
pdf |
| 99 |  | Rbpj1(?)/Panc1-Rbpj1-ChIP-Seq(GSE47459)/Homer | 1e-2 | -5.168e+00 | 0.0142 | 1439.0 | 15.02% | 1038.9 | 14.12% | motif file (matrix) |
pdf |
| 100 |  | Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.053e+00 | 0.0157 | 177.0 | 1.85% | 112.1 | 1.52% | motif file (matrix) |
pdf |
| 101 |  | E2A-nearPU.1(HLH)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-2 | -4.922e+00 | 0.0177 | 1460.0 | 15.24% | 1056.6 | 14.36% | motif file (matrix) |
pdf |
| 102 |  | EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer | 1e-2 | -4.778e+00 | 0.0203 | 616.0 | 6.43% | 430.9 | 5.86% | motif file (matrix) |
pdf |
| 103 |  | Tcf12(HLH)/GM12878-Tcf12-ChIP-Seq(GSE32465)/Homer | 1e-2 | -4.756e+00 | 0.0205 | 1102.0 | 11.50% | 790.8 | 10.75% | motif file (matrix) |
pdf |
| 104 |  | CEBP:CEBP(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer | 1e-2 | -4.752e+00 | 0.0205 | 84.0 | 0.88% | 49.4 | 0.67% | motif file (matrix) |
pdf |
| 105 |  | Nkx3.1(Homeobox)/LNCaP-Nkx3.1-ChIP-Seq(GSE28264)/Homer | 1e-2 | -4.741e+00 | 0.0205 | 2089.0 | 21.81% | 1531.5 | 20.82% | motif file (matrix) |
pdf |

















































































































































































































