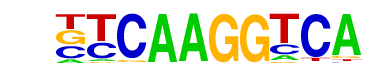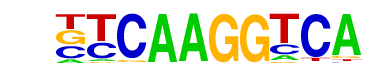| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | BORIS(Zf)/K562-CTCFL-ChIP-Seq(GSE32465)/Homer | 1e-130 | -3.002e+02 | 0.0000 | 390.0 | 10.24% | 52.2 | 2.28% | motif file (matrix) |
pdf |
| 2 |  | CTCF(Zf)/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-129 | -2.992e+02 | 0.0000 | 304.0 | 7.98% | 31.7 | 1.38% | motif file (matrix) |
pdf |
| 3 |  | FOXA1(Forkhead)/MCF7-FOXA1-ChIP-Seq(GSE26831)/Homer | 1e-129 | -2.974e+02 | 0.0000 | 549.0 | 14.41% | 100.6 | 4.39% | motif file (matrix) |
pdf |
| 4 |  | Gata1(Zf)/K562-GATA1-ChIP-Seq(GSE18829)/Homer | 1e-124 | -2.858e+02 | 0.0000 | 451.0 | 11.84% | 72.7 | 3.17% | motif file (matrix) |
pdf |
| 5 |  | Gata2(Zf)/K562-GATA2-ChIP-Seq(GSE18829)/Homer | 1e-122 | -2.817e+02 | 0.0000 | 484.0 | 12.71% | 83.5 | 3.64% | motif file (matrix) |
pdf |
| 6 |  | FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-117 | -2.697e+02 | 0.0000 | 639.0 | 16.78% | 139.2 | 6.08% | motif file (matrix) |
pdf |
| 7 |  | Sp1(Zf)/Promoter/Homer | 1e-115 | -2.658e+02 | 0.0000 | 545.0 | 14.31% | 107.8 | 4.71% | motif file (matrix) |
pdf |
| 8 |  | Gata4(Zf)/Heart-Gata4-ChIP-Seq(GSE35151)/Homer | 1e-114 | -2.627e+02 | 0.0000 | 641.0 | 16.83% | 142.2 | 6.21% | motif file (matrix) |
pdf |
| 9 |  | GATA3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-112 | -2.581e+02 | 0.0000 | 782.0 | 20.53% | 198.7 | 8.67% | motif file (matrix) |
pdf |
| 10 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-105 | -2.430e+02 | 0.0000 | 529.0 | 13.89% | 108.3 | 4.73% | motif file (matrix) |
pdf |
| 11 |  | Foxa2(Forkhead)/Liver-Foxa2-ChIP-Seq(GSE25694)/Homer | 1e-89 | -2.059e+02 | 0.0000 | 505.0 | 13.26% | 111.0 | 4.85% | motif file (matrix) |
pdf |
| 12 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-75 | -1.737e+02 | 0.0000 | 509.0 | 13.36% | 124.7 | 5.45% | motif file (matrix) |
pdf |
| 13 |  | KLF5(Zf)/LoVo-KLF5-ChIP-Seq(GSE49402)/Homer | 1e-72 | -1.665e+02 | 0.0000 | 1091.0 | 28.64% | 386.2 | 16.86% | motif file (matrix) |
pdf |
| 14 |  | Sox3(HMG)/NPC-Sox3-ChIP-Seq(GSE33059)/Homer | 1e-71 | -1.654e+02 | 0.0000 | 831.0 | 21.82% | 264.7 | 11.56% | motif file (matrix) |
pdf |
| 15 |  | Fox:Ebox(Forkhead:HLH)/Panc1-Foxa2-ChIP-Seq(GSE47459)/Homer | 1e-70 | -1.623e+02 | 0.0000 | 585.0 | 15.36% | 159.5 | 6.96% | motif file (matrix) |
pdf |
| 16 |  | ETS(ETS)/Promoter/Homer | 1e-65 | -1.501e+02 | 0.0000 | 342.0 | 8.98% | 71.7 | 3.13% | motif file (matrix) |
pdf |
| 17 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-62 | -1.435e+02 | 0.0000 | 304.0 | 7.98% | 61.0 | 2.66% | motif file (matrix) |
pdf |
| 18 |  | Sox2(HMG)/mES-Sox2-ChIP-Seq(GSE11431)/Homer | 1e-62 | -1.435e+02 | 0.0000 | 490.0 | 12.86% | 129.1 | 5.64% | motif file (matrix) |
pdf |
| 19 |  | Foxo1(Forkhead)/RAW-Foxo1-ChIP-Seq(Fan et al.)/Homer | 1e-61 | -1.407e+02 | 0.0000 | 887.0 | 23.29% | 306.8 | 13.39% | motif file (matrix) |
pdf |
| 20 | 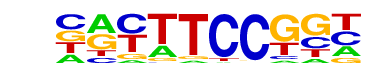 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-59 | -1.371e+02 | 0.0000 | 762.0 | 20.01% | 251.0 | 10.96% | motif file (matrix) |
pdf |
| 21 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-58 | -1.358e+02 | 0.0000 | 479.0 | 12.58% | 128.4 | 5.60% | motif file (matrix) |
pdf |
| 22 |  | Hnf1(Homeobox)/Liver-Foxa2-Chip-Seq(GSE25694)/Homer | 1e-57 | -1.324e+02 | 0.0000 | 117.0 | 3.07% | 10.9 | 0.48% | motif file (matrix) |
pdf |
| 23 | 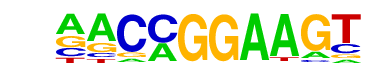 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-56 | -1.296e+02 | 0.0000 | 883.0 | 23.18% | 312.6 | 13.65% | motif file (matrix) |
pdf |
| 24 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-56 | -1.296e+02 | 0.0000 | 659.0 | 17.30% | 208.6 | 9.11% | motif file (matrix) |
pdf |
| 25 |  | Sox6(HMG)/Myotubes-Sox6-ChIP-Seq(GSE32627)/Homer | 1e-55 | -1.270e+02 | 0.0000 | 715.0 | 18.77% | 235.4 | 10.28% | motif file (matrix) |
pdf |
| 26 |  | Unknown-ESC-element/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-53 | -1.242e+02 | 0.0000 | 446.0 | 11.71% | 120.6 | 5.27% | motif file (matrix) |
pdf |
| 27 |  | Rfx2(HTH)/LoVo-RFX2-ChIP-Seq(GSE49402)/Homer | 1e-53 | -1.240e+02 | 0.0000 | 91.0 | 2.39% | 6.1 | 0.27% | motif file (matrix) |
pdf |
| 28 | 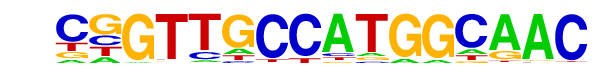 | RFX(HTH)/K562-RFX3-ChIP-Seq(SRA012198)/Homer | 1e-50 | -1.174e+02 | 0.0000 | 88.0 | 2.31% | 6.6 | 0.29% | motif file (matrix) |
pdf |
| 29 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-50 | -1.163e+02 | 0.0000 | 387.0 | 10.16% | 100.3 | 4.38% | motif file (matrix) |
pdf |
| 30 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-49 | -1.129e+02 | 0.0000 | 673.0 | 17.67% | 225.1 | 9.83% | motif file (matrix) |
pdf |
| 31 |  | TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer | 1e-47 | -1.096e+02 | 0.0000 | 407.0 | 10.69% | 111.5 | 4.87% | motif file (matrix) |
pdf |
| 32 |  | NFY(CCAAT)/Promoter/Homer | 1e-45 | -1.040e+02 | 0.0000 | 479.0 | 12.58% | 144.2 | 6.29% | motif file (matrix) |
pdf |
| 33 |  | HNF4a(NR/DR1)/HepG2-HNF4a-ChIP-Seq(GSE25021)/Homer | 1e-41 | -9.645e+01 | 0.0000 | 352.0 | 9.24% | 95.4 | 4.17% | motif file (matrix) |
pdf |
| 34 |  | TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer | 1e-39 | -9.102e+01 | 0.0000 | 290.0 | 7.61% | 73.0 | 3.19% | motif file (matrix) |
pdf |
| 35 |  | Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-37 | -8.701e+01 | 0.0000 | 142.0 | 3.73% | 23.3 | 1.02% | motif file (matrix) |
pdf |
| 36 |  | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-37 | -8.646e+01 | 0.0000 | 657.0 | 17.25% | 237.9 | 10.39% | motif file (matrix) |
pdf |
| 37 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-37 | -8.608e+01 | 0.0000 | 282.0 | 7.40% | 72.8 | 3.18% | motif file (matrix) |
pdf |
| 38 |  | SPDEF(ETS)/VCaP-SPDEF-ChIP-Seq(SRA014231)/Homer | 1e-37 | -8.589e+01 | 0.0000 | 530.0 | 13.91% | 178.6 | 7.80% | motif file (matrix) |
pdf |
| 39 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-34 | -8.032e+01 | 0.0000 | 273.0 | 7.17% | 71.4 | 3.12% | motif file (matrix) |
pdf |
| 40 |  | Nanog(Homeobox)/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-33 | -7.817e+01 | 0.0000 | 1503.0 | 39.46% | 690.8 | 30.16% | motif file (matrix) |
pdf |
| 41 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-33 | -7.610e+01 | 0.0000 | 899.0 | 23.60% | 367.8 | 16.06% | motif file (matrix) |
pdf |
| 42 |  | TR4(NR/DR1)/Hela-TR4-ChIP-Seq(GSE24685)/Homer | 1e-27 | -6.421e+01 | 0.0000 | 121.0 | 3.18% | 22.2 | 0.97% | motif file (matrix) |
pdf |
| 43 |  | Nkx6.1(Homeobox)/Islet-Nkx6.1-ChIP-Seq(GSE40975)/Homer | 1e-26 | -6.199e+01 | 0.0000 | 799.0 | 20.98% | 332.0 | 14.49% | motif file (matrix) |
pdf |
| 44 |  | CRE(bZIP)/Promoter/Homer | 1e-25 | -5.866e+01 | 0.0000 | 146.0 | 3.83% | 32.9 | 1.44% | motif file (matrix) |
pdf |
| 45 |  | Usf2(HLH)/C2C12-Usf2-ChIP-Seq(GSE36030)/Homer | 1e-24 | -5.653e+01 | 0.0000 | 187.0 | 4.91% | 48.8 | 2.13% | motif file (matrix) |
pdf |
| 46 |  | Erra(NR)/HepG2-Erra-ChIP-Seq(GSE31477)/Homer | 1e-24 | -5.579e+01 | 0.0000 | 1025.0 | 26.91% | 458.1 | 20.00% | motif file (matrix) |
pdf |
| 47 |  | Rfx5(HTH)/GM12878-Rfx5-ChIP-Seq(GSE31477)/Homer | 1e-24 | -5.546e+01 | 0.0000 | 178.0 | 4.67% | 45.8 | 2.00% | motif file (matrix) |
pdf |
| 48 |  | AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-22 | -5.080e+01 | 0.0000 | 292.0 | 7.67% | 95.8 | 4.18% | motif file (matrix) |
pdf |
| 49 |  | E-box(HLH)/Promoter/Homer | 1e-21 | -4.999e+01 | 0.0000 | 66.0 | 1.73% | 10.0 | 0.43% | motif file (matrix) |
pdf |
| 50 |  | X-box(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-21 | -4.893e+01 | 0.0000 | 69.0 | 1.81% | 10.5 | 0.46% | motif file (matrix) |
pdf |
| 51 |  | EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer | 1e-21 | -4.873e+01 | 0.0000 | 300.0 | 7.88% | 100.4 | 4.38% | motif file (matrix) |
pdf |
| 52 |  | GFY-Staf/Promoters/Homer | 1e-20 | -4.748e+01 | 0.0000 | 68.0 | 1.79% | 10.1 | 0.44% | motif file (matrix) |
pdf |
| 53 |  | Tcf3(HMG)/mES-Tcf3-ChIP-Seq(GSE11724)/Homer | 1e-20 | -4.697e+01 | 0.0000 | 145.0 | 3.81% | 36.9 | 1.61% | motif file (matrix) |
pdf |
| 54 |  | USF1(HLH)/GM12878-Usf1-ChIP-Seq(GSE32465)/Homer | 1e-19 | -4.582e+01 | 0.0000 | 232.0 | 6.09% | 72.1 | 3.15% | motif file (matrix) |
pdf |
| 55 |  | bHLHE40(HLH)/HepG2-BHLHE40-ChIP-Seq(GSE31477)/Homer | 1e-18 | -4.304e+01 | 0.0000 | 156.0 | 4.10% | 42.9 | 1.87% | motif file (matrix) |
pdf |
| 56 |  | c-Jun-CRE(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-18 | -4.265e+01 | 0.0000 | 168.0 | 4.41% | 47.6 | 2.08% | motif file (matrix) |
pdf |
| 57 |  | Tcf4(HMG)/Hct116-Tcf4-ChIP-Seq(SRA012054)/Homer | 1e-18 | -4.230e+01 | 0.0000 | 213.0 | 5.59% | 66.9 | 2.92% | motif file (matrix) |
pdf |
| 58 |  | Lhx3(Homeobox)/Neuron-Lhx3-ChIP-Seq(GSE31456)/Homer | 1e-17 | -3.995e+01 | 0.0000 | 522.0 | 13.70% | 216.0 | 9.43% | motif file (matrix) |
pdf |
| 59 |  | Maz(Zf)/HepG2-Maz-ChIP-Seq(GSE31477)/Homer | 1e-17 | -3.990e+01 | 0.0000 | 970.0 | 25.47% | 452.8 | 19.77% | motif file (matrix) |
pdf |
| 60 |  | CLOCK(HLH)/Liver-Clock-ChIP-Seq(GSE39860)/Homer | 1e-16 | -3.911e+01 | 0.0000 | 249.0 | 6.54% | 84.3 | 3.68% | motif file (matrix) |
pdf |
| 61 |  | CRX(Homeobox)/Retina-Crx-ChIP-Seq(GSE20012)/Homer | 1e-16 | -3.834e+01 | 0.0000 | 816.0 | 21.42% | 371.4 | 16.22% | motif file (matrix) |
pdf |
| 62 |  | Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer | 1e-16 | -3.815e+01 | 0.0000 | 150.0 | 3.94% | 42.7 | 1.86% | motif file (matrix) |
pdf |
| 63 |  | Eomes(T-box)/H9-Eomes-ChIP-Seq(GSE26097)/Homer | 1e-16 | -3.685e+01 | 0.0000 | 752.0 | 19.74% | 339.7 | 14.83% | motif file (matrix) |
pdf |
| 64 |  | NRF1(NRF)/MCF7-NRF1-ChIP-Seq(Unpublished)/Homer | 1e-15 | -3.603e+01 | 0.0000 | 157.0 | 4.12% | 47.0 | 2.05% | motif file (matrix) |
pdf |
| 65 |  | SCL(HLH)/HPC7-Scl-ChIP-Seq(GSE13511)/Homer | 1e-15 | -3.585e+01 | 0.0000 | 1985.0 | 52.11% | 1043.7 | 45.56% | motif file (matrix) |
pdf |
| 66 |  | GATA:SCL/Ter119-SCL-ChIP-Seq(GSE18720)/Homer | 1e-15 | -3.559e+01 | 0.0000 | 81.0 | 2.13% | 17.9 | 0.78% | motif file (matrix) |
pdf |
| 67 |  | Bach2(bZIP)/OCILy7-Bach2-ChIP-Seq(GSE44420)/Homer | 1e-15 | -3.462e+01 | 0.0000 | 105.0 | 2.76% | 26.9 | 1.17% | motif file (matrix) |
pdf |
| 68 |  | REST-NRSF(Zf)/Jurkat-NRSF-ChIP-Seq/Homer | 1e-14 | -3.378e+01 | 0.0000 | 20.0 | 0.53% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 69 |  | Lhx2(Homeobox)/HFSC-Lhx2-ChIP-Seq(GSE48068)/Homer | 1e-14 | -3.346e+01 | 0.0000 | 318.0 | 8.35% | 121.4 | 5.30% | motif file (matrix) |
pdf |
| 70 |  | ELF5(ETS)/T47D-ELF5-ChIP-Seq(GSE30407)/Homer | 1e-13 | -3.144e+01 | 0.0000 | 383.0 | 10.06% | 155.8 | 6.80% | motif file (matrix) |
pdf |
| 71 |  | Isl1(Homeobox)/Neuron-Isl1-ChIP-Seq(GSE31456)/Homer | 1e-13 | -3.129e+01 | 0.0000 | 627.0 | 16.46% | 281.9 | 12.31% | motif file (matrix) |
pdf |
| 72 |  | GFY(?)/Promoter/Homer | 1e-13 | -3.000e+01 | 0.0000 | 58.0 | 1.52% | 11.2 | 0.49% | motif file (matrix) |
pdf |
| 73 |  | Atf1(bZIP)/K562-ATF1-ChIP-Seq(GSE31477)/Homer | 1e-12 | -2.959e+01 | 0.0000 | 279.0 | 7.32% | 107.0 | 4.67% | motif file (matrix) |
pdf |
| 74 |  | RUNX2(Runt)/PCa-RUNX2-ChIP-Seq(GSE33889)/Homer | 1e-12 | -2.922e+01 | 0.0000 | 301.0 | 7.90% | 117.1 | 5.11% | motif file (matrix) |
pdf |
| 75 |  | Klf4(Zf)/mES-Klf4-ChIP-Seq(GSE11431)/Homer | 1e-12 | -2.849e+01 | 0.0000 | 332.0 | 8.72% | 133.6 | 5.83% | motif file (matrix) |
pdf |
| 76 |  | NPAS2(HLH)/Liver-NPAS2-ChIP-Seq(GSE39860)/Homer | 1e-12 | -2.836e+01 | 0.0000 | 429.0 | 11.26% | 182.7 | 7.98% | motif file (matrix) |
pdf |
| 77 |  | RUNX-AML(Runt)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer | 1e-11 | -2.688e+01 | 0.0000 | 261.0 | 6.85% | 100.6 | 4.39% | motif file (matrix) |
pdf |
| 78 |  | JunD(bZIP)/K562-JunD-ChIP-Seq/Homer | 1e-11 | -2.574e+01 | 0.0000 | 63.0 | 1.65% | 15.0 | 0.65% | motif file (matrix) |
pdf |
| 79 |  | Rfx1(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-11 | -2.553e+01 | 0.0000 | 114.0 | 2.99% | 34.4 | 1.50% | motif file (matrix) |
pdf |
| 80 |  | Hoxb4(Homeobox)/ES-Hoxb4-ChIP-Seq(GSE34014)/Homer | 1e-10 | -2.518e+01 | 0.0000 | 68.0 | 1.79% | 16.7 | 0.73% | motif file (matrix) |
pdf |
| 81 |  | NF1:FOXA1/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-10 | -2.431e+01 | 0.0000 | 26.0 | 0.68% | 3.4 | 0.15% | motif file (matrix) |
pdf |
| 82 |  | Oct2(POU/Homeobox)/Bcell-Oct2-ChIP-Seq(GSE21512)/Homer | 1e-10 | -2.422e+01 | 0.0000 | 75.0 | 1.97% | 19.4 | 0.84% | motif file (matrix) |
pdf |
| 83 |  | Oct4:Sox17/F9-Sox17-ChIP-Seq(GSE44553)/Homer | 1e-10 | -2.330e+01 | 0.0000 | 43.0 | 1.13% | 8.4 | 0.37% | motif file (matrix) |
pdf |
| 84 |  | ARE(NR)/LNCAP-AR-ChIP-Seq(GSE27824)/Homer | 1e-9 | -2.275e+01 | 0.0000 | 86.0 | 2.26% | 24.6 | 1.07% | motif file (matrix) |
pdf |
| 85 |  | Max(HLH)/K562-Max-ChIP-Seq(GSE31477)/Homer | 1e-9 | -2.131e+01 | 0.0000 | 257.0 | 6.75% | 104.1 | 4.55% | motif file (matrix) |
pdf |
| 86 |  | MafA(bZIP)/Islet-MafA-ChIP-Seq(GSE30298)/Homer | 1e-9 | -2.104e+01 | 0.0000 | 357.0 | 9.37% | 155.3 | 6.78% | motif file (matrix) |
pdf |
| 87 |  | NFkB-p65(RHD)/GM12787-p65-ChIP-Seq(GSE19485)/Homer | 1e-9 | -2.085e+01 | 0.0000 | 184.0 | 4.83% | 69.9 | 3.05% | motif file (matrix) |
pdf |
| 88 |  | YY1(Zf)/Promoter/Homer | 1e-9 | -2.076e+01 | 0.0000 | 71.0 | 1.86% | 19.2 | 0.84% | motif file (matrix) |
pdf |
| 89 |  | Pdx1(Homeobox)/Islet-Pdx1-ChIP-Seq(SRA008281)/Homer | 1e-8 | -2.055e+01 | 0.0000 | 235.0 | 6.17% | 94.2 | 4.11% | motif file (matrix) |
pdf |
| 90 |  | Oct4(POU/Homeobox)/mES-Oct4-ChIP-Seq(GSE11431)/Homer | 1e-8 | -2.054e+01 | 0.0000 | 116.0 | 3.05% | 38.8 | 1.69% | motif file (matrix) |
pdf |
| 91 |  | Znf263(Zf)/K562-Znf263-ChIP-Seq(GSE31477)/Homer | 1e-8 | -1.905e+01 | 0.0000 | 972.0 | 25.52% | 495.8 | 21.65% | motif file (matrix) |
pdf |
| 92 |  | RUNX(Runt)/HPC7-Runx1-ChIP-Seq(GSE22178)/Homer | 1e-8 | -1.851e+01 | 0.0000 | 274.0 | 7.19% | 116.9 | 5.10% | motif file (matrix) |
pdf |
| 93 |  | BMAL1(HLH)/Liver-Bmal1-ChIP-Seq(GSE39860)/Homer | 1e-7 | -1.818e+01 | 0.0000 | 566.0 | 14.86% | 271.6 | 11.86% | motif file (matrix) |
pdf |
| 94 |  | RXR(NR/DR1)/3T3L1-RXR-ChIP-Seq(GSE13511)/Homer | 1e-7 | -1.711e+01 | 0.0000 | 593.0 | 15.57% | 288.1 | 12.58% | motif file (matrix) |
pdf |
| 95 |  | Stat3(Stat)/mES-Stat3-ChIP-Seq(GSE11431)/Homer | 1e-7 | -1.681e+01 | 0.0000 | 238.0 | 6.25% | 100.0 | 4.37% | motif file (matrix) |
pdf |
| 96 |  | Tbox:Smad/ESCd5-Smad2_3-ChIP-Seq(GSE29422)/Homer | 1e-7 | -1.677e+01 | 0.0000 | 85.0 | 2.23% | 27.3 | 1.19% | motif file (matrix) |
pdf |
| 97 |  | ZNF143|STAF(Zf)/CUTLL-ZNF143-ChIP-Seq(GSE29600)/Homer | 1e-7 | -1.652e+01 | 0.0000 | 179.0 | 4.70% | 71.2 | 3.11% | motif file (matrix) |
pdf |
| 98 |  | Ap4(HLH)/AML-Tfap4-ChIP-Seq(GSE45738)/Homer | 1e-7 | -1.635e+01 | 0.0000 | 590.0 | 15.49% | 288.1 | 12.58% | motif file (matrix) |
pdf |
| 99 |  | TCFL2(HMG)/K562-TCF7L2-ChIP-Seq(GSE29196)/Homer | 1e-7 | -1.634e+01 | 0.0000 | 53.0 | 1.39% | 14.6 | 0.64% | motif file (matrix) |
pdf |
| 100 |  | Ptf1a(HLH)/Panc1-Ptf1a-ChIP-Seq(GSE47459)/Homer | 1e-6 | -1.605e+01 | 0.0000 | 1162.0 | 30.51% | 612.9 | 26.76% | motif file (matrix) |
pdf |
| 101 |  | Tcf12(HLH)/GM12878-Tcf12-ChIP-Seq(GSE32465)/Homer | 1e-6 | -1.577e+01 | 0.0000 | 511.0 | 13.42% | 246.8 | 10.77% | motif file (matrix) |
pdf |
| 102 |  | CTCF-SatelliteElement/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-6 | -1.542e+01 | 0.0000 | 12.0 | 0.32% | 1.0 | 0.04% | motif file (matrix) |
pdf |
| 103 |  | NRF1/Promoter/Homer | 1e-6 | -1.448e+01 | 0.0000 | 156.0 | 4.10% | 62.1 | 2.71% | motif file (matrix) |
pdf |
| 104 |  | Esrrb(NR)/mES-Esrrb-ChIP-Seq(GSE11431)/Homer | 1e-6 | -1.430e+01 | 0.0000 | 243.0 | 6.38% | 106.1 | 4.63% | motif file (matrix) |
pdf |
| 105 |  | PPARE(NR/DR1)/3T3L1-Pparg-ChIP-Seq(GSE13511)/Homer | 1e-5 | -1.373e+01 | 0.0000 | 512.0 | 13.44% | 251.3 | 10.97% | motif file (matrix) |
pdf |
| 106 |  | NFAT:AP1/Jurkat-NFATC1-ChIP-Seq(Jolma et al.)/Homer | 1e-5 | -1.346e+01 | 0.0000 | 47.0 | 1.23% | 13.7 | 0.60% | motif file (matrix) |
pdf |
| 107 |  | STAT4(Stat)/CD4-Stat4-ChIP-Seq(GSE22104)/Homer | 1e-5 | -1.277e+01 | 0.0000 | 337.0 | 8.85% | 158.8 | 6.93% | motif file (matrix) |
pdf |
| 108 |  | HOXA9(Homeobox)/HSC-Hoxa9-ChIP-Seq(GSE33509)/Homer | 1e-5 | -1.235e+01 | 0.0000 | 149.0 | 3.91% | 62.0 | 2.71% | motif file (matrix) |
pdf |
| 109 |  | Bcl6(Zf)/Liver-Bcl6-ChIP-Seq(GSE31578)/Homer | 1e-5 | -1.231e+01 | 0.0000 | 486.0 | 12.76% | 240.8 | 10.51% | motif file (matrix) |
pdf |
| 110 |  | E2F(E2F)/Cell-Cycle-Exp/Homer | 1e-5 | -1.183e+01 | 0.0000 | 40.0 | 1.05% | 11.7 | 0.51% | motif file (matrix) |
pdf |
| 111 | 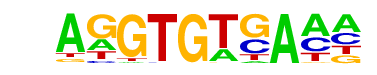 | Tbet(T-box)/CD8-Tbet-ChIP-Seq(GSE33802)/Homer | 1e-4 | -1.087e+01 | 0.0000 | 365.0 | 9.58% | 178.0 | 7.77% | motif file (matrix) |
pdf |
| 112 |  | Nrf2(bZIP)/Lymphoblast-Nrf2-ChIP-Seq(GSE37589)/Homer | 1e-4 | -1.082e+01 | 0.0000 | 23.0 | 0.60% | 5.2 | 0.23% | motif file (matrix) |
pdf |
| 113 |  | Stat3+il21(Stat)/CD4-Stat3-ChIP-Seq(GSE19198)/Homer | 1e-4 | -1.050e+01 | 0.0001 | 284.0 | 7.46% | 134.1 | 5.86% | motif file (matrix) |
pdf |
| 114 |  | RUNX1(Runt)/Jurkat-RUNX1-ChIP-Seq(GSE29180)/Homer | 1e-4 | -1.047e+01 | 0.0001 | 367.0 | 9.64% | 179.4 | 7.83% | motif file (matrix) |
pdf |
| 115 | 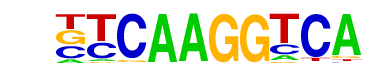 | Nr5a2(NR)/Pancreas-LRH1-ChIP-Seq(GSE34295)/Homer | 1e-4 | -1.023e+01 | 0.0001 | 238.0 | 6.25% | 110.2 | 4.81% | motif file (matrix) |
pdf |
| 116 |  | PRDM14(Zf)/H1-PRDM14-ChIP-Seq(GSE22767)/Homer | 1e-4 | -9.998e+00 | 0.0001 | 107.0 | 2.81% | 43.8 | 1.91% | motif file (matrix) |
pdf |
| 117 |  | c-Myc(HLH)/LNCAP-cMyc-ChIP-Seq(unpublished)/Homer | 1e-4 | -9.873e+00 | 0.0001 | 233.0 | 6.12% | 108.8 | 4.75% | motif file (matrix) |
pdf |
| 118 |  | Nur77(NR)/K562-NR4A1-ChIP-Seq(GSE31363)/Homer | 1e-4 | -9.816e+00 | 0.0001 | 86.0 | 2.26% | 33.5 | 1.46% | motif file (matrix) |
pdf |
| 119 |  | NeuroD1(bHLH)/Islet-NeuroD1-ChIP-Seq(GSE30298)/Homer | 1e-4 | -9.608e+00 | 0.0001 | 387.0 | 10.16% | 192.2 | 8.39% | motif file (matrix) |
pdf |
| 120 |  | GRE/RAW264.7-GRE-ChIP-Seq(Unpublished)/Homer | 1e-4 | -9.511e+00 | 0.0002 | 62.0 | 1.63% | 23.0 | 1.00% | motif file (matrix) |
pdf |
| 121 |  | GATA-DR4(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-4 | -9.431e+00 | 0.0002 | 27.0 | 0.71% | 7.0 | 0.31% | motif file (matrix) |
pdf |
| 122 |  | CEBP:AP1(bZIP)/ThioMac-CEBPb-ChIP-Seq(GSE21512)/Homer | 1e-4 | -9.355e+00 | 0.0002 | 180.0 | 4.73% | 81.7 | 3.57% | motif file (matrix) |
pdf |
| 123 |  | Nkx2.1(Homeobox)/LungAC-Nkx2.1-ChIP-Seq(GSE43252)/Homer | 1e-4 | -9.322e+00 | 0.0002 | 979.0 | 25.70% | 529.6 | 23.12% | motif file (matrix) |
pdf |
| 124 |  | GATA-IR4(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-3 | -9.082e+00 | 0.0002 | 24.0 | 0.63% | 6.6 | 0.29% | motif file (matrix) |
pdf |
| 125 |  | ERE(NR/IR3)/MCF7-ERa-ChIP-Seq(Unpublished)/Homer | 1e-3 | -8.808e+00 | 0.0003 | 90.0 | 2.36% | 36.6 | 1.60% | motif file (matrix) |
pdf |
| 126 |  | GRE(NR/IR3)/A549-GR-ChIP-Seq(GSE32465)/Homer | 1e-3 | -8.755e+00 | 0.0003 | 36.0 | 0.95% | 11.5 | 0.50% | motif file (matrix) |
pdf |
| 127 |  | GATA-IR3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-3 | -8.516e+00 | 0.0004 | 38.0 | 1.00% | 12.3 | 0.54% | motif file (matrix) |
pdf |
| 128 |  | E2F4(E2F)/K562-E2F4-ChIP-Seq(GSE31477)/Homer | 1e-3 | -8.251e+00 | 0.0005 | 310.0 | 8.14% | 153.3 | 6.69% | motif file (matrix) |
pdf |
| 129 |  | NF-E2(bZIP)/K562-NFE2-ChIP-Seq(GSE31477)/Homer | 1e-3 | -8.175e+00 | 0.0005 | 23.0 | 0.60% | 6.6 | 0.29% | motif file (matrix) |
pdf |
| 130 |  | Pbx3(Homeobox)/GM12878-PBX3-ChIP-Seq(GSE32465)/Homer | 1e-3 | -8.001e+00 | 0.0006 | 114.0 | 2.99% | 49.6 | 2.17% | motif file (matrix) |
pdf |
| 131 |  | Atoh1(bHLH)/Cerebellum-Atoh1-ChIP-Seq(GSE22111)/Homer | 1e-3 | -7.980e+00 | 0.0006 | 499.0 | 13.10% | 259.6 | 11.33% | motif file (matrix) |
pdf |
| 132 |  | PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-3 | -7.926e+00 | 0.0007 | 440.0 | 11.55% | 226.6 | 9.89% | motif file (matrix) |
pdf |
| 133 |  | MyoG(HLH)/C2C12-MyoG-ChIP-Seq(GSE36024)/Homer | 1e-3 | -7.868e+00 | 0.0007 | 532.0 | 13.97% | 278.4 | 12.15% | motif file (matrix) |
pdf |
| 134 |  | AP-2gamma(AP2)/MCF7-TFAP2C-ChIP-Seq(GSE21234)/Homer | 1e-3 | -7.797e+00 | 0.0008 | 551.0 | 14.47% | 289.7 | 12.65% | motif file (matrix) |
pdf |
| 135 |  | PAX3:FKHR-fusion(Paired/Homeobox)/Rh4-PAX3:FKHR-ChIP-Seq(GSE19063)/Homer | 1e-3 | -7.522e+00 | 0.0010 | 41.0 | 1.08% | 14.7 | 0.64% | motif file (matrix) |
pdf |
| 136 |  | TRa(NR)/C17.2-TRa-ChIP-Seq(GSE38347)/Homer | 1e-3 | -7.474e+00 | 0.0010 | 170.0 | 4.46% | 79.3 | 3.46% | motif file (matrix) |
pdf |
| 137 |  | FOXA1:AR/LNCAP-AR-ChIP-Seq(GSE27824)/Homer | 1e-2 | -6.907e+00 | 0.0018 | 19.0 | 0.50% | 5.2 | 0.23% | motif file (matrix) |
pdf |
| 138 |  | n-Myc(HLH)/mES-nMyc-ChIP-Seq(GSE11431)/Homer | 1e-2 | -6.786e+00 | 0.0020 | 267.0 | 7.01% | 133.2 | 5.82% | motif file (matrix) |
pdf |
| 139 |  | Pax7(Paired/Homeobox)/Myoblast-Pax7-ChIP-Seq(GSE25064)/Homer | 1e-2 | -6.484e+00 | 0.0027 | 21.0 | 0.55% | 6.9 | 0.30% | motif file (matrix) |
pdf |
| 140 |  | p53(p53)/mES-cMyc-ChIP-Seq(GSE11431)/Homer | 1e-2 | -6.404e+00 | 0.0029 | 7.0 | 0.18% | 1.8 | 0.08% | motif file (matrix) |
pdf |
| 141 |  | PRDM9(Zf)/Testis-DMC1-ChIP-Seq(GSE35498)/Homer | 1e-2 | -6.253e+00 | 0.0034 | 145.0 | 3.81% | 68.8 | 3.00% | motif file (matrix) |
pdf |
| 142 |  | HOXA2(Homeobox)/mES-Hoxa2-ChIP-Seq(Donaldson et al.)/Homer | 1e-2 | -6.174e+00 | 0.0036 | 23.0 | 0.60% | 7.6 | 0.33% | motif file (matrix) |
pdf |
| 143 |  | MyoD(HLH)/Myotube-MyoD-ChIP-Seq(GSE21614)/Homer | 1e-2 | -5.709e+00 | 0.0057 | 399.0 | 10.48% | 210.6 | 9.20% | motif file (matrix) |
pdf |
| 144 |  | GATA-DR8(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-2 | -5.232e+00 | 0.0091 | 17.0 | 0.45% | 5.5 | 0.24% | motif file (matrix) |
pdf |
| 145 |  | Tlx?(NR)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-2 | -4.894e+00 | 0.0127 | 191.0 | 5.01% | 96.5 | 4.21% | motif file (matrix) |
pdf |
| 146 |  | Tbx5(T-box)/HL1-Tbx5.biotin-ChIP-Seq(GSE21529)/Homer | 1e-2 | -4.726e+00 | 0.0149 | 1295.0 | 34.00% | 737.7 | 32.21% | motif file (matrix) |
pdf |
| 147 |  | ETS:RUNX/Jurkat-RUNX1-ChIP-Seq(GSE17954)/Homer | 1e-2 | -4.723e+00 | 0.0149 | 58.0 | 1.52% | 25.1 | 1.09% | motif file (matrix) |
pdf |
| 148 |  | Unknown(Homeobox)/Limb-p300-ChIP-Seq/Homer | 1e-2 | -4.705e+00 | 0.0150 | 128.0 | 3.36% | 62.4 | 2.72% | motif file (matrix) |
pdf |
| 149 |  | Smad3(MAD)/NPC-Smad3-ChIP-Seq(GSE36673)/Homer | 1e-2 | -4.664e+00 | 0.0156 | 1193.0 | 31.32% | 677.5 | 29.58% | motif file (matrix) |
pdf |