| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-351 | -8.083e+02 | 0.0000 | 1309.0 | 23.74% | 2958.2 | 6.73% | motif file (matrix) |
pdf |
| 2 |  | FOXA1(Forkhead)/MCF7-FOXA1-ChIP-Seq(GSE26831)/Homer | 1e-339 | -7.827e+02 | 0.0000 | 1126.0 | 20.42% | 2271.3 | 5.16% | motif file (matrix) |
pdf |
| 3 |  | Foxa2(Forkhead)/Liver-Foxa2-ChIP-Seq(GSE25694)/Homer | 1e-289 | -6.657e+02 | 0.0000 | 1023.0 | 18.56% | 2167.0 | 4.93% | motif file (matrix) |
pdf |
| 4 |  | Fox:Ebox(Forkhead:HLH)/Panc1-Foxa2-ChIP-Seq(GSE47459)/Homer | 1e-216 | -4.994e+02 | 0.0000 | 1154.0 | 20.93% | 3320.0 | 7.55% | motif file (matrix) |
pdf |
| 5 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-152 | -3.507e+02 | 0.0000 | 536.0 | 9.72% | 1094.3 | 2.49% | motif file (matrix) |
pdf |
| 6 |  | CTCF(Zf)/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-138 | -3.181e+02 | 0.0000 | 377.0 | 6.84% | 600.6 | 1.37% | motif file (matrix) |
pdf |
| 7 |  | BORIS(Zf)/K562-CTCFL-ChIP-Seq(GSE32465)/Homer | 1e-126 | -2.917e+02 | 0.0000 | 494.0 | 8.96% | 1095.2 | 2.49% | motif file (matrix) |
pdf |
| 8 |  | Foxo1(Forkhead)/RAW-Foxo1-ChIP-Seq(Fan et al.)/Homer | 1e-94 | -2.176e+02 | 0.0000 | 1446.0 | 26.23% | 6763.6 | 15.38% | motif file (matrix) |
pdf |
| 9 |  | Gata4(Zf)/Heart-Gata4-ChIP-Seq(GSE35151)/Homer | 1e-76 | -1.770e+02 | 0.0000 | 710.0 | 12.88% | 2664.4 | 6.06% | motif file (matrix) |
pdf |
| 10 |  | KLF5(Zf)/LoVo-KLF5-ChIP-Seq(GSE49402)/Homer | 1e-76 | -1.755e+02 | 0.0000 | 1574.0 | 28.55% | 8049.1 | 18.30% | motif file (matrix) |
pdf |
| 11 |  | GATA3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-71 | -1.653e+02 | 0.0000 | 924.0 | 16.76% | 3991.0 | 9.08% | motif file (matrix) |
pdf |
| 12 |  | Sp1(Zf)/Promoter/Homer | 1e-71 | -1.647e+02 | 0.0000 | 627.0 | 11.37% | 2290.0 | 5.21% | motif file (matrix) |
pdf |
| 13 |  | Gata2(Zf)/K562-GATA2-ChIP-Seq(GSE18829)/Homer | 1e-67 | -1.564e+02 | 0.0000 | 508.0 | 9.21% | 1710.5 | 3.89% | motif file (matrix) |
pdf |
| 14 |  | Lhx3(Homeobox)/Neuron-Lhx3-ChIP-Seq(GSE31456)/Homer | 1e-64 | -1.486e+02 | 0.0000 | 983.0 | 17.83% | 4502.8 | 10.24% | motif file (matrix) |
pdf |
| 15 |  | Unknown-ESC-element/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-63 | -1.460e+02 | 0.0000 | 725.0 | 13.15% | 2973.5 | 6.76% | motif file (matrix) |
pdf |
| 16 |  | Gata1(Zf)/K562-GATA1-ChIP-Seq(GSE18829)/Homer | 1e-58 | -1.357e+02 | 0.0000 | 457.0 | 8.29% | 1564.7 | 3.56% | motif file (matrix) |
pdf |
| 17 |  | Eomes(T-box)/H9-Eomes-ChIP-Seq(GSE26097)/Homer | 1e-50 | -1.172e+02 | 0.0000 | 1350.0 | 24.49% | 7265.5 | 16.52% | motif file (matrix) |
pdf |
| 18 |  | Nkx6.1(Homeobox)/Islet-Nkx6.1-ChIP-Seq(GSE40975)/Homer | 1e-48 | -1.106e+02 | 0.0000 | 1285.0 | 23.31% | 6914.5 | 15.72% | motif file (matrix) |
pdf |
| 19 |  | Lhx2(Homeobox)/HFSC-Lhx2-ChIP-Seq(GSE48068)/Homer | 1e-46 | -1.073e+02 | 0.0000 | 626.0 | 11.35% | 2714.0 | 6.17% | motif file (matrix) |
pdf |
| 20 |  | NFY(CCAAT)/Promoter/Homer | 1e-44 | -1.033e+02 | 0.0000 | 683.0 | 12.39% | 3094.9 | 7.04% | motif file (matrix) |
pdf |
| 21 |  | Rfx2(HTH)/LoVo-RFX2-ChIP-Seq(GSE49402)/Homer | 1e-40 | -9.428e+01 | 0.0000 | 178.0 | 3.23% | 426.5 | 0.97% | motif file (matrix) |
pdf |
| 22 |  | Sox3(HMG)/NPC-Sox3-ChIP-Seq(GSE33059)/Homer | 1e-39 | -9.119e+01 | 0.0000 | 1093.0 | 19.83% | 5886.7 | 13.39% | motif file (matrix) |
pdf |
| 23 |  | Maz(Zf)/HepG2-Maz-ChIP-Seq(GSE31477)/Homer | 1e-39 | -9.030e+01 | 0.0000 | 1574.0 | 28.55% | 9249.0 | 21.03% | motif file (matrix) |
pdf |
| 24 | 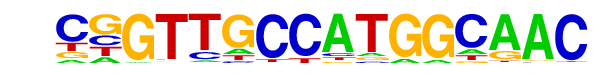 | RFX(HTH)/K562-RFX3-ChIP-Seq(SRA012198)/Homer | 1e-37 | -8.745e+01 | 0.0000 | 165.0 | 2.99% | 395.2 | 0.90% | motif file (matrix) |
pdf |
| 25 |  | Klf4(Zf)/mES-Klf4-ChIP-Seq(GSE11431)/Homer | 1e-37 | -8.533e+01 | 0.0000 | 499.0 | 9.05% | 2158.7 | 4.91% | motif file (matrix) |
pdf |
| 26 | 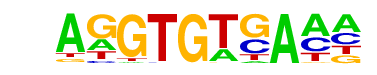 | Tbet(T-box)/CD8-Tbet-ChIP-Seq(GSE33802)/Homer | 1e-36 | -8.451e+01 | 0.0000 | 699.0 | 12.68% | 3391.8 | 7.71% | motif file (matrix) |
pdf |
| 27 |  | Tbx5(T-box)/HL1-Tbx5.biotin-ChIP-Seq(GSE21529)/Homer | 1e-33 | -7.789e+01 | 0.0000 | 2161.0 | 39.20% | 13814.6 | 31.41% | motif file (matrix) |
pdf |
| 28 |  | X-box(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-33 | -7.664e+01 | 0.0000 | 156.0 | 2.83% | 395.2 | 0.90% | motif file (matrix) |
pdf |
| 29 |  | Sox2(HMG)/mES-Sox2-ChIP-Seq(GSE11431)/Homer | 1e-32 | -7.417e+01 | 0.0000 | 599.0 | 10.87% | 2879.9 | 6.55% | motif file (matrix) |
pdf |
| 30 |  | Nanog(Homeobox)/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-32 | -7.393e+01 | 0.0000 | 2284.0 | 41.43% | 14836.9 | 33.74% | motif file (matrix) |
pdf |
| 31 |  | Isl1(Homeobox)/Neuron-Isl1-ChIP-Seq(GSE31456)/Homer | 1e-31 | -7.174e+01 | 0.0000 | 1138.0 | 20.64% | 6498.0 | 14.78% | motif file (matrix) |
pdf |
| 32 |  | Tcf4(HMG)/Hct116-Tcf4-ChIP-Seq(SRA012054)/Homer | 1e-30 | -7.119e+01 | 0.0000 | 325.0 | 5.90% | 1275.2 | 2.90% | motif file (matrix) |
pdf |
| 33 |  | Oct4(POU/Homeobox)/mES-Oct4-ChIP-Seq(GSE11431)/Homer | 1e-29 | -6.775e+01 | 0.0000 | 248.0 | 4.50% | 882.6 | 2.01% | motif file (matrix) |
pdf |
| 34 |  | Rfx1(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-28 | -6.541e+01 | 0.0000 | 227.0 | 4.12% | 787.7 | 1.79% | motif file (matrix) |
pdf |
| 35 |  | TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer | 1e-27 | -6.269e+01 | 0.0000 | 548.0 | 9.94% | 2691.2 | 6.12% | motif file (matrix) |
pdf |
| 36 |  | CRX(Homeobox)/Retina-Crx-ChIP-Seq(GSE20012)/Homer | 1e-27 | -6.257e+01 | 0.0000 | 1317.0 | 23.89% | 7929.6 | 18.03% | motif file (matrix) |
pdf |
| 37 |  | Sox6(HMG)/Myotubes-Sox6-ChIP-Seq(GSE32627)/Homer | 1e-25 | -5.947e+01 | 0.0000 | 903.0 | 16.38% | 5084.7 | 11.56% | motif file (matrix) |
pdf |
| 38 |  | Oct2(POU/Homeobox)/Bcell-Oct2-ChIP-Seq(GSE21512)/Homer | 1e-23 | -5.508e+01 | 0.0000 | 160.0 | 2.90% | 507.5 | 1.15% | motif file (matrix) |
pdf |
| 39 |  | Tcf3(HMG)/mES-Tcf3-ChIP-Seq(GSE11724)/Homer | 1e-23 | -5.383e+01 | 0.0000 | 201.0 | 3.65% | 723.8 | 1.65% | motif file (matrix) |
pdf |
| 40 |  | TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer | 1e-21 | -4.946e+01 | 0.0000 | 382.0 | 6.93% | 1807.9 | 4.11% | motif file (matrix) |
pdf |
| 41 |  | Rfx5(HTH)/GM12878-Rfx5-ChIP-Seq(GSE31477)/Homer | 1e-21 | -4.862e+01 | 0.0000 | 293.0 | 5.31% | 1279.1 | 2.91% | motif file (matrix) |
pdf |
| 42 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-18 | -4.181e+01 | 0.0000 | 504.0 | 9.14% | 2684.2 | 6.10% | motif file (matrix) |
pdf |
| 43 |  | SCL(HLH)/HPC7-Scl-ChIP-Seq(GSE13511)/Homer | 1e-17 | -4.124e+01 | 0.0000 | 3009.0 | 54.58% | 21413.9 | 48.69% | motif file (matrix) |
pdf |
| 44 |  | CRE(bZIP)/Promoter/Homer | 1e-16 | -3.904e+01 | 0.0000 | 200.0 | 3.63% | 825.7 | 1.88% | motif file (matrix) |
pdf |
| 45 |  | Ptf1a(HLH)/Panc1-Ptf1a-ChIP-Seq(GSE47459)/Homer | 1e-15 | -3.638e+01 | 0.0000 | 1882.0 | 34.14% | 12778.7 | 29.06% | motif file (matrix) |
pdf |
| 46 |  | ETS(ETS)/Promoter/Homer | 1e-15 | -3.621e+01 | 0.0000 | 332.0 | 6.02% | 1651.3 | 3.76% | motif file (matrix) |
pdf |
| 47 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-15 | -3.543e+01 | 0.0000 | 519.0 | 9.41% | 2885.4 | 6.56% | motif file (matrix) |
pdf |
| 48 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-15 | -3.488e+01 | 0.0000 | 520.0 | 9.43% | 2901.2 | 6.60% | motif file (matrix) |
pdf |
| 49 |  | E2A(HLH)/proBcell-E2A-ChIP-Seq(GSE21978)/Homer | 1e-15 | -3.462e+01 | 0.0000 | 1206.0 | 21.88% | 7769.9 | 17.67% | motif file (matrix) |
pdf |
| 50 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-14 | -3.313e+01 | 0.0000 | 718.0 | 13.02% | 4297.2 | 9.77% | motif file (matrix) |
pdf |
| 51 |  | c-Jun-CRE(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-13 | -3.159e+01 | 0.0000 | 200.0 | 3.63% | 892.7 | 2.03% | motif file (matrix) |
pdf |
| 52 |  | Rbpj1(?)/Panc1-Rbpj1-ChIP-Seq(GSE47459)/Homer | 1e-13 | -3.089e+01 | 0.0000 | 913.0 | 16.56% | 5736.8 | 13.05% | motif file (matrix) |
pdf |
| 53 | 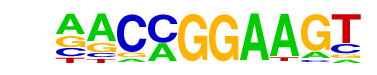 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-13 | -3.052e+01 | 0.0000 | 1003.0 | 18.19% | 6397.2 | 14.55% | motif file (matrix) |
pdf |
| 54 |  | Pitx1(Homeobox)/Chicken-Pitx1-ChIP-Seq(GSE38910)/Homer | 1e-12 | -2.968e+01 | 0.0000 | 2102.0 | 38.13% | 14698.5 | 33.42% | motif file (matrix) |
pdf |
| 55 |  | Tcf12(HLH)/GM12878-Tcf12-ChIP-Seq(GSE32465)/Homer | 1e-12 | -2.915e+01 | 0.0000 | 857.0 | 15.55% | 5377.5 | 12.23% | motif file (matrix) |
pdf |
| 56 | 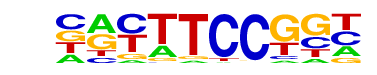 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-12 | -2.769e+01 | 0.0000 | 834.0 | 15.13% | 5249.0 | 11.94% | motif file (matrix) |
pdf |
| 57 | 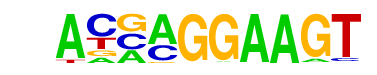 | ELF5(ETS)/T47D-ELF5-ChIP-Seq(GSE30407)/Homer | 1e-11 | -2.616e+01 | 0.0000 | 494.0 | 8.96% | 2885.0 | 6.56% | motif file (matrix) |
pdf |
| 58 |  | E-box(HLH)/Promoter/Homer | 1e-11 | -2.535e+01 | 0.0000 | 85.0 | 1.54% | 296.8 | 0.67% | motif file (matrix) |
pdf |
| 59 |  | Ap4(HLH)/AML-Tfap4-ChIP-Seq(GSE45738)/Homer | 1e-10 | -2.532e+01 | 0.0000 | 954.0 | 17.30% | 6185.4 | 14.07% | motif file (matrix) |
pdf |
| 60 |  | CTCF-SatelliteElement/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-10 | -2.492e+01 | 0.0000 | 28.0 | 0.51% | 45.4 | 0.10% | motif file (matrix) |
pdf |
| 61 |  | EKLF(Zf)/Erythrocyte-Klf1-ChIP-Seq(GSE20478)/Homer | 1e-10 | -2.453e+01 | 0.0000 | 206.0 | 3.74% | 1005.7 | 2.29% | motif file (matrix) |
pdf |
| 62 |  | Usf2(HLH)/C2C12-Usf2-ChIP-Seq(GSE36030)/Homer | 1e-10 | -2.394e+01 | 0.0000 | 196.0 | 3.56% | 950.3 | 2.16% | motif file (matrix) |
pdf |
| 63 |  | Atf1(bZIP)/K562-ATF1-ChIP-Seq(GSE31477)/Homer | 1e-10 | -2.338e+01 | 0.0000 | 359.0 | 6.51% | 2018.9 | 4.59% | motif file (matrix) |
pdf |
| 64 |  | GFY-Staf/Promoters/Homer | 1e-10 | -2.311e+01 | 0.0000 | 64.0 | 1.16% | 203.7 | 0.46% | motif file (matrix) |
pdf |
| 65 |  | OCT4-SOX2-TCF-NANOG((POU/Homeobox/HMG)/mES-Oct4-ChIP-Seq(GSE11431)/Homer | 1e-9 | -2.291e+01 | 0.0000 | 73.0 | 1.32% | 249.2 | 0.57% | motif file (matrix) |
pdf |
| 66 |  | FOXA1:AR/LNCAP-AR-ChIP-Seq(GSE27824)/Homer | 1e-9 | -2.284e+01 | 0.0000 | 39.0 | 0.71% | 91.2 | 0.21% | motif file (matrix) |
pdf |
| 67 |  | TCFL2(HMG)/K562-TCF7L2-ChIP-Seq(GSE29196)/Homer | 1e-9 | -2.253e+01 | 0.0000 | 70.0 | 1.27% | 236.9 | 0.54% | motif file (matrix) |
pdf |
| 68 |  | REST-NRSF(Zf)/Jurkat-NRSF-ChIP-Seq/Homer | 1e-9 | -2.219e+01 | 0.0000 | 26.0 | 0.47% | 45.0 | 0.10% | motif file (matrix) |
pdf |
| 69 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-9 | -2.209e+01 | 0.0000 | 725.0 | 13.15% | 4613.8 | 10.49% | motif file (matrix) |
pdf |
| 70 |  | JunD(bZIP)/K562-JunD-ChIP-Seq/Homer | 1e-9 | -2.180e+01 | 0.0000 | 79.0 | 1.43% | 287.1 | 0.65% | motif file (matrix) |
pdf |
| 71 |  | SPDEF(ETS)/VCaP-SPDEF-ChIP-Seq(SRA014231)/Homer | 1e-9 | -2.093e+01 | 0.0000 | 678.0 | 12.30% | 4306.1 | 9.79% | motif file (matrix) |
pdf |
| 72 |  | Phox2a(Homeobox)/Neuron-Phox2a-ChIP-Seq(GSE31456)/Homer | 1e-9 | -2.087e+01 | 0.0000 | 136.0 | 2.47% | 618.9 | 1.41% | motif file (matrix) |
pdf |
| 73 |  | NRF1(NRF)/MCF7-NRF1-ChIP-Seq(Unpublished)/Homer | 1e-9 | -2.082e+01 | 0.0000 | 183.0 | 3.32% | 907.4 | 2.06% | motif file (matrix) |
pdf |
| 74 |  | NRF1/Promoter/Homer | 1e-8 | -1.943e+01 | 0.0000 | 197.0 | 3.57% | 1014.3 | 2.31% | motif file (matrix) |
pdf |
| 75 |  | HNF6(Homeobox)/Liver-Hnf6-ChIP-Seq(ERP000394)/Homer | 1e-8 | -1.942e+01 | 0.0000 | 221.0 | 4.01% | 1169.6 | 2.66% | motif file (matrix) |
pdf |
| 76 |  | Myf5(bHLH)/GM-Myf5-ChIP-Seq(GSE24852)/Homer | 1e-8 | -1.936e+01 | 0.0000 | 572.0 | 10.38% | 3588.5 | 8.16% | motif file (matrix) |
pdf |
| 77 |  | Pbx3(Homeobox)/GM12878-PBX3-ChIP-Seq(GSE32465)/Homer | 1e-7 | -1.837e+01 | 0.0000 | 195.0 | 3.54% | 1016.3 | 2.31% | motif file (matrix) |
pdf |
| 78 |  | MyoG(HLH)/C2C12-MyoG-ChIP-Seq(GSE36024)/Homer | 1e-7 | -1.800e+01 | 0.0000 | 855.0 | 15.51% | 5687.7 | 12.93% | motif file (matrix) |
pdf |
| 79 |  | NeuroD1(bHLH)/Islet-NeuroD1-ChIP-Seq(GSE30298)/Homer | 1e-7 | -1.760e+01 | 0.0000 | 568.0 | 10.30% | 3607.1 | 8.20% | motif file (matrix) |
pdf |
| 80 |  | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-7 | -1.758e+01 | 0.0000 | 786.0 | 14.26% | 5193.9 | 11.81% | motif file (matrix) |
pdf |
| 81 |  | GFY(?)/Promoter/Homer | 1e-7 | -1.744e+01 | 0.0000 | 71.0 | 1.29% | 274.3 | 0.62% | motif file (matrix) |
pdf |
| 82 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-7 | -1.738e+01 | 0.0000 | 1034.0 | 18.76% | 7040.6 | 16.01% | motif file (matrix) |
pdf |
| 83 |  | GATA:SCL/Ter119-SCL-ChIP-Seq(GSE18720)/Homer | 1e-7 | -1.713e+01 | 0.0000 | 87.0 | 1.58% | 366.9 | 0.83% | motif file (matrix) |
pdf |
| 84 |  | Oct4:Sox17/F9-Sox17-ChIP-Seq(GSE44553)/Homer | 1e-7 | -1.677e+01 | 0.0000 | 73.0 | 1.32% | 290.3 | 0.66% | motif file (matrix) |
pdf |
| 85 |  | Znf263(Zf)/K562-Znf263-ChIP-Seq(GSE31477)/Homer | 1e-7 | -1.654e+01 | 0.0000 | 1407.0 | 25.52% | 9895.3 | 22.50% | motif file (matrix) |
pdf |
| 86 |  | bHLHE40(HLH)/HepG2-BHLHE40-ChIP-Seq(GSE31477)/Homer | 1e-6 | -1.494e+01 | 0.0000 | 192.0 | 3.48% | 1049.4 | 2.39% | motif file (matrix) |
pdf |
| 87 |  | PBX1(Homeobox)/MCF7-PBX1-ChIP-Seq(GSE28007)/Homer | 1e-6 | -1.444e+01 | 0.0000 | 85.0 | 1.54% | 379.9 | 0.86% | motif file (matrix) |
pdf |
| 88 |  | USF1(HLH)/GM12878-Usf1-ChIP-Seq(GSE32465)/Homer | 1e-6 | -1.428e+01 | 0.0000 | 260.0 | 4.72% | 1519.9 | 3.46% | motif file (matrix) |
pdf |
| 89 |  | E2A-nearPU.1(HLH)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-6 | -1.409e+01 | 0.0000 | 1003.0 | 18.19% | 6940.0 | 15.78% | motif file (matrix) |
pdf |
| 90 |  | Olig2(bHLH)/Neuron-Olig2-ChIP-Seq(GSE30882)/Homer | 1e-5 | -1.287e+01 | 0.0000 | 1101.0 | 19.97% | 7734.1 | 17.59% | motif file (matrix) |
pdf |
| 91 |  | E2F4(E2F)/K562-E2F4-ChIP-Seq(GSE31477)/Homer | 1e-5 | -1.220e+01 | 0.0000 | 382.0 | 6.93% | 2425.4 | 5.52% | motif file (matrix) |
pdf |
| 92 |  | PU.1(ETS)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-5 | -1.198e+01 | 0.0000 | 308.0 | 5.59% | 1905.8 | 4.33% | motif file (matrix) |
pdf |
| 93 |  | CLOCK(HLH)/Liver-Clock-ChIP-Seq(GSE39860)/Homer | 1e-5 | -1.151e+01 | 0.0000 | 295.0 | 5.35% | 1825.2 | 4.15% | motif file (matrix) |
pdf |
| 94 |  | Six1(Homeobox)/Myoblast-Six1-ChIP-Chip(GSE20150)/Homer | 1e-4 | -1.095e+01 | 0.0000 | 124.0 | 2.25% | 666.4 | 1.52% | motif file (matrix) |
pdf |
| 95 |  | Nur77(NR)/K562-NR4A1-ChIP-Seq(GSE31363)/Homer | 1e-4 | -1.053e+01 | 0.0001 | 98.0 | 1.78% | 504.1 | 1.15% | motif file (matrix) |
pdf |
| 96 |  | Atoh1(bHLH)/Cerebellum-Atoh1-ChIP-Seq(GSE22111)/Homer | 1e-4 | -1.045e+01 | 0.0001 | 740.0 | 13.42% | 5120.1 | 11.64% | motif file (matrix) |
pdf |
| 97 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-4 | -1.018e+01 | 0.0001 | 275.0 | 4.99% | 1718.1 | 3.91% | motif file (matrix) |
pdf |
| 98 |  | Egr1(Zf)/K562-Egr1-ChIP-Seq(GSE32465)/Homer | 1e-4 | -1.012e+01 | 0.0001 | 568.0 | 10.30% | 3851.8 | 8.76% | motif file (matrix) |
pdf |
| 99 |  | Foxh1(Forkhead)/hESC-FOXH1-ChIP-Seq(GSE29422)/Homer | 1e-4 | -1.009e+01 | 0.0001 | 268.0 | 4.86% | 1671.5 | 3.80% | motif file (matrix) |
pdf |
| 100 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-4 | -9.830e+00 | 0.0001 | 271.0 | 4.92% | 1699.1 | 3.86% | motif file (matrix) |
pdf |
| 101 |  | c-Myc(HLH)/LNCAP-cMyc-ChIP-Seq(unpublished)/Homer | 1e-4 | -9.809e+00 | 0.0001 | 292.0 | 5.30% | 1850.0 | 4.21% | motif file (matrix) |
pdf |
| 102 |  | EBF1(EBF)/Near-E2A-ChIP-Seq(GSE21512)/Homer | 1e-4 | -9.611e+00 | 0.0002 | 806.0 | 14.62% | 5655.1 | 12.86% | motif file (matrix) |
pdf |
| 103 |  | IRF4(IRF)/GM12878-IRF4-ChIP-Seq(GSE32465)/Homer | 1e-4 | -9.251e+00 | 0.0002 | 195.0 | 3.54% | 1179.2 | 2.68% | motif file (matrix) |
pdf |
| 104 |  | MyoD(HLH)/Myotube-MyoD-ChIP-Seq(GSE21614)/Homer | 1e-3 | -8.947e+00 | 0.0003 | 628.0 | 11.39% | 4347.3 | 9.89% | motif file (matrix) |
pdf |
| 105 |  | Egr2/Thymocytes-Egr2-ChIP-Seq(GSE34254)/Homer | 1e-3 | -8.747e+00 | 0.0004 | 152.0 | 2.76% | 893.0 | 2.03% | motif file (matrix) |
pdf |
| 106 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-3 | -8.567e+00 | 0.0004 | 379.0 | 6.87% | 2517.4 | 5.72% | motif file (matrix) |
pdf |
| 107 |  | Hnf1(Homeobox)/Liver-Foxa2-Chip-Seq(GSE25694)/Homer | 1e-3 | -8.426e+00 | 0.0005 | 42.0 | 0.76% | 183.3 | 0.42% | motif file (matrix) |
pdf |
| 108 |  | E2F6(E2F)/Hela-E2F6-ChIP-Seq(GSE31477)/Homer | 1e-3 | -8.381e+00 | 0.0005 | 429.0 | 7.78% | 2891.6 | 6.58% | motif file (matrix) |
pdf |
| 109 |  | Tbx20(T-box)/Heart-Tbx20-ChIP-Seq(GSE29636)/Homer | 1e-3 | -8.271e+00 | 0.0006 | 144.0 | 2.61% | 848.3 | 1.93% | motif file (matrix) |
pdf |
| 110 |  | NF1:FOXA1/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-3 | -8.245e+00 | 0.0006 | 33.0 | 0.60% | 133.3 | 0.30% | motif file (matrix) |
pdf |
| 111 |  | ZNF143|STAF(Zf)/CUTLL-ZNF143-ChIP-Seq(GSE29600)/Homer | 1e-3 | -8.098e+00 | 0.0007 | 233.0 | 4.23% | 1476.4 | 3.36% | motif file (matrix) |
pdf |
| 112 |  | n-Myc(HLH)/mES-nMyc-ChIP-Seq(GSE11431)/Homer | 1e-3 | -7.614e+00 | 0.0011 | 357.0 | 6.48% | 2390.8 | 5.44% | motif file (matrix) |
pdf |
| 113 |  | Tbox:Smad/ESCd5-Smad2_3-ChIP-Seq(GSE29422)/Homer | 1e-3 | -7.496e+00 | 0.0012 | 118.0 | 2.14% | 686.9 | 1.56% | motif file (matrix) |
pdf |
| 114 |  | MafA(bZIP)/Islet-MafA-ChIP-Seq(GSE30298)/Homer | 1e-3 | -7.107e+00 | 0.0018 | 486.0 | 8.82% | 3367.6 | 7.66% | motif file (matrix) |
pdf |
| 115 |  | Srebp2(HLH)/HepG2-Srebp2-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.029e+00 | 0.0019 | 92.0 | 1.67% | 519.2 | 1.18% | motif file (matrix) |
pdf |
| 116 |  | Nkx3.1(Homeobox)/LNCaP-Nkx3.1-ChIP-Seq(GSE28264)/Homer | 1e-2 | -6.810e+00 | 0.0023 | 1170.0 | 21.22% | 8603.1 | 19.56% | motif file (matrix) |
pdf |
| 117 |  | Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer | 1e-2 | -6.449e+00 | 0.0033 | 179.0 | 3.25% | 1137.2 | 2.59% | motif file (matrix) |
pdf |
| 118 |  | Max(HLH)/K562-Max-ChIP-Seq(GSE31477)/Homer | 1e-2 | -6.243e+00 | 0.0041 | 313.0 | 5.68% | 2118.7 | 4.82% | motif file (matrix) |
pdf |
| 119 |  | PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-2 | -6.220e+00 | 0.0041 | 655.0 | 11.88% | 4687.6 | 10.66% | motif file (matrix) |
pdf |
| 120 |  | Stat3(Stat)/mES-Stat3-ChIP-Seq(GSE11431)/Homer | 1e-2 | -5.848e+00 | 0.0059 | 277.0 | 5.02% | 1868.5 | 4.25% | motif file (matrix) |
pdf |
| 121 |  | GATA-DR4(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-2 | -5.387e+00 | 0.0093 | 36.0 | 0.65% | 178.8 | 0.41% | motif file (matrix) |
pdf |
| 122 |  | Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.357e+00 | 0.0095 | 105.0 | 1.90% | 642.9 | 1.46% | motif file (matrix) |
pdf |
| 123 |  | E2F1(E2F)/Hela-E2F1-ChIP-Seq(GSE22478)/Homer | 1e-2 | -5.306e+00 | 0.0099 | 198.0 | 3.59% | 1309.2 | 2.98% | motif file (matrix) |
pdf |
| 124 |  | Smad3(MAD)/NPC-Smad3-ChIP-Seq(GSE36673)/Homer | 1e-2 | -5.090e+00 | 0.0122 | 1708.0 | 30.98% | 12942.8 | 29.43% | motif file (matrix) |
pdf |
| 125 |  | ETS:RUNX/Jurkat-RUNX1-ChIP-Seq(GSE17954)/Homer | 1e-2 | -5.060e+00 | 0.0125 | 68.0 | 1.23% | 393.5 | 0.89% | motif file (matrix) |
pdf |
| 126 |  | MafK(bZIP)/C2C12-MafK-ChIP-Seq(GSE36030)/Homer | 1e-2 | -5.051e+00 | 0.0125 | 127.0 | 2.30% | 805.1 | 1.83% | motif file (matrix) |
pdf |
| 127 |  | EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer | 1e-2 | -4.834e+00 | 0.0154 | 347.0 | 6.29% | 2432.2 | 5.53% | motif file (matrix) |
pdf |
| 128 |  | GATA-IR3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-2 | -4.816e+00 | 0.0156 | 51.0 | 0.93% | 283.2 | 0.64% | motif file (matrix) |
pdf |
| 129 |  | STAT6/Macrophage-Stat6-ChIP-Seq(GSE38377)/Homer | 1e-2 | -4.685e+00 | 0.0176 | 226.0 | 4.10% | 1537.6 | 3.50% | motif file (matrix) |
pdf |

































































































































































































































































