| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-107 | -2.468e+02 | 0.0000 | 691.0 | 21.77% | 4111.1 | 8.84% | motif file (matrix) |
pdf |
| 2 |  | FOXA1(Forkhead)/MCF7-FOXA1-ChIP-Seq(GSE26831)/Homer | 1e-99 | -2.290e+02 | 0.0000 | 580.0 | 18.27% | 3223.4 | 6.93% | motif file (matrix) |
pdf |
| 3 |  | Foxa2(Forkhead)/Liver-Foxa2-ChIP-Seq(GSE25694)/Homer | 1e-91 | -2.114e+02 | 0.0000 | 534.0 | 16.82% | 2949.6 | 6.34% | motif file (matrix) |
pdf |
| 4 |  | CTCF(Zf)/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-63 | -1.460e+02 | 0.0000 | 162.0 | 5.10% | 444.1 | 0.95% | motif file (matrix) |
pdf |
| 5 |  | BORIS(Zf)/K562-CTCFL-ChIP-Seq(GSE32465)/Homer | 1e-56 | -1.299e+02 | 0.0000 | 187.0 | 5.89% | 664.8 | 1.43% | motif file (matrix) |
pdf |
| 6 |  | Fox:Ebox(Forkhead:HLH)/Panc1-Foxa2-ChIP-Seq(GSE47459)/Homer | 1e-52 | -1.212e+02 | 0.0000 | 581.0 | 18.30% | 4389.4 | 9.43% | motif file (matrix) |
pdf |
| 7 |  | Unknown-ESC-element/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-48 | -1.112e+02 | 0.0000 | 425.0 | 13.39% | 2886.4 | 6.20% | motif file (matrix) |
pdf |
| 8 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-45 | -1.057e+02 | 0.0000 | 272.0 | 8.57% | 1488.1 | 3.20% | motif file (matrix) |
pdf |
| 9 |  | Eomes(T-box)/H9-Eomes-ChIP-Seq(GSE26097)/Homer | 1e-37 | -8.659e+01 | 0.0000 | 946.0 | 29.80% | 9372.5 | 20.14% | motif file (matrix) |
pdf |
| 10 |  | Lhx2(Homeobox)/HFSC-Lhx2-ChIP-Seq(GSE48068)/Homer | 1e-27 | -6.394e+01 | 0.0000 | 447.0 | 14.08% | 3821.0 | 8.21% | motif file (matrix) |
pdf |
| 11 |  | Tbx5(T-box)/HL1-Tbx5.biotin-ChIP-Seq(GSE21529)/Homer | 1e-25 | -5.919e+01 | 0.0000 | 1353.0 | 42.63% | 15625.2 | 33.58% | motif file (matrix) |
pdf |
| 12 |  | Lhx3(Homeobox)/Neuron-Lhx3-ChIP-Seq(GSE31456)/Homer | 1e-23 | -5.487e+01 | 0.0000 | 621.0 | 19.57% | 6093.8 | 13.10% | motif file (matrix) |
pdf |
| 13 |  | Gata4(Zf)/Heart-Gata4-ChIP-Seq(GSE35151)/Homer | 1e-23 | -5.317e+01 | 0.0000 | 428.0 | 13.48% | 3815.0 | 8.20% | motif file (matrix) |
pdf |
| 14 |  | CRX(Homeobox)/Retina-Crx-ChIP-Seq(GSE20012)/Homer | 1e-22 | -5.205e+01 | 0.0000 | 927.0 | 29.21% | 10094.6 | 21.70% | motif file (matrix) |
pdf |
| 15 |  | Gata2(Zf)/K562-GATA2-ChIP-Seq(GSE18829)/Homer | 1e-22 | -5.186e+01 | 0.0000 | 305.0 | 9.61% | 2449.2 | 5.26% | motif file (matrix) |
pdf |
| 16 |  | Gata1(Zf)/K562-GATA1-ChIP-Seq(GSE18829)/Homer | 1e-19 | -4.594e+01 | 0.0000 | 273.0 | 8.60% | 2199.1 | 4.73% | motif file (matrix) |
pdf |
| 17 |  | Foxo1(Forkhead)/RAW-Foxo1-ChIP-Seq(Fan et al.)/Homer | 1e-19 | -4.575e+01 | 0.0000 | 772.0 | 24.32% | 8271.9 | 17.78% | motif file (matrix) |
pdf |
| 18 | 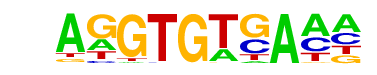 | Tbet(T-box)/CD8-Tbet-ChIP-Seq(GSE33802)/Homer | 1e-18 | -4.372e+01 | 0.0000 | 475.0 | 14.97% | 4589.9 | 9.87% | motif file (matrix) |
pdf |
| 19 |  | GATA3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-17 | -4.104e+01 | 0.0000 | 566.0 | 17.83% | 5786.7 | 12.44% | motif file (matrix) |
pdf |
| 20 |  | Nkx6.1(Homeobox)/Islet-Nkx6.1-ChIP-Seq(GSE40975)/Homer | 1e-17 | -4.023e+01 | 0.0000 | 878.0 | 27.66% | 9858.9 | 21.19% | motif file (matrix) |
pdf |
| 21 |  | Sox3(HMG)/NPC-Sox3-ChIP-Seq(GSE33059)/Homer | 1e-13 | -3.050e+01 | 0.0000 | 650.0 | 20.48% | 7215.4 | 15.51% | motif file (matrix) |
pdf |
| 22 |  | Sox6(HMG)/Myotubes-Sox6-ChIP-Seq(GSE32627)/Homer | 1e-12 | -2.848e+01 | 0.0000 | 566.0 | 17.83% | 6197.4 | 13.32% | motif file (matrix) |
pdf |
| 23 |  | Pitx1(Homeobox)/Chicken-Pitx1-ChIP-Seq(GSE38910)/Homer | 1e-10 | -2.487e+01 | 0.0000 | 1427.0 | 44.96% | 18214.9 | 39.15% | motif file (matrix) |
pdf |
| 24 |  | Phox2a(Homeobox)/Neuron-Phox2a-ChIP-Seq(GSE31456)/Homer | 1e-10 | -2.475e+01 | 0.0000 | 131.0 | 4.13% | 1019.1 | 2.19% | motif file (matrix) |
pdf |
| 25 |  | Oct4(POU/Homeobox)/mES-Oct4-ChIP-Seq(GSE11431)/Homer | 1e-10 | -2.312e+01 | 0.0000 | 153.0 | 4.82% | 1286.2 | 2.76% | motif file (matrix) |
pdf |
| 26 |  | Nanog(Homeobox)/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-9 | -2.256e+01 | 0.0000 | 1416.0 | 44.61% | 18196.2 | 39.11% | motif file (matrix) |
pdf |
| 27 |  | Sox2(HMG)/mES-Sox2-ChIP-Seq(GSE11431)/Homer | 1e-9 | -2.222e+01 | 0.0000 | 323.0 | 10.18% | 3325.9 | 7.15% | motif file (matrix) |
pdf |
| 28 |  | Isl1(Homeobox)/Neuron-Isl1-ChIP-Seq(GSE31456)/Homer | 1e-9 | -2.183e+01 | 0.0000 | 709.0 | 22.34% | 8371.5 | 17.99% | motif file (matrix) |
pdf |
| 29 |  | Ptf1a(HLH)/Panc1-Ptf1a-ChIP-Seq(GSE47459)/Homer | 1e-9 | -2.083e+01 | 0.0000 | 1050.0 | 33.08% | 13114.2 | 28.19% | motif file (matrix) |
pdf |
| 30 |  | Tcf4(HMG)/Hct116-Tcf4-ChIP-Seq(SRA012054)/Homer | 1e-8 | -1.969e+01 | 0.0000 | 180.0 | 5.67% | 1666.7 | 3.58% | motif file (matrix) |
pdf |
| 31 |  | E2A-nearPU.1(HLH)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-8 | -1.924e+01 | 0.0000 | 599.0 | 18.87% | 7020.3 | 15.09% | motif file (matrix) |
pdf |
| 32 |  | E2A(HLH)/proBcell-E2A-ChIP-Seq(GSE21978)/Homer | 1e-7 | -1.835e+01 | 0.0000 | 613.0 | 19.31% | 7252.2 | 15.59% | motif file (matrix) |
pdf |
| 33 |  | SCL(HLH)/HPC7-Scl-ChIP-Seq(GSE13511)/Homer | 1e-7 | -1.650e+01 | 0.0000 | 1674.0 | 52.74% | 22355.6 | 48.05% | motif file (matrix) |
pdf |
| 34 |  | Oct2(POU/Homeobox)/Bcell-Oct2-ChIP-Seq(GSE21512)/Homer | 1e-7 | -1.640e+01 | 0.0000 | 91.0 | 2.87% | 729.3 | 1.57% | motif file (matrix) |
pdf |
| 35 |  | OCT4-SOX2-TCF-NANOG((POU/Homeobox/HMG)/mES-Oct4-ChIP-Seq(GSE11431)/Homer | 1e-7 | -1.621e+01 | 0.0000 | 60.0 | 1.89% | 411.4 | 0.88% | motif file (matrix) |
pdf |
| 36 |  | Tcf3(HMG)/mES-Tcf3-ChIP-Seq(GSE11724)/Homer | 1e-6 | -1.604e+01 | 0.0000 | 109.0 | 3.43% | 933.0 | 2.01% | motif file (matrix) |
pdf |
| 37 |  | KLF5(Zf)/LoVo-KLF5-ChIP-Seq(GSE49402)/Homer | 1e-6 | -1.593e+01 | 0.0000 | 486.0 | 15.31% | 5675.8 | 12.20% | motif file (matrix) |
pdf |
| 38 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-5 | -1.380e+01 | 0.0000 | 605.0 | 19.06% | 7392.2 | 15.89% | motif file (matrix) |
pdf |
| 39 |  | HNF6(Homeobox)/Liver-Hnf6-ChIP-Seq(ERP000394)/Homer | 1e-5 | -1.362e+01 | 0.0000 | 165.0 | 5.20% | 1646.7 | 3.54% | motif file (matrix) |
pdf |
| 40 |  | Znf263(Zf)/K562-Znf263-ChIP-Seq(GSE31477)/Homer | 1e-5 | -1.228e+01 | 0.0000 | 698.0 | 21.99% | 8765.1 | 18.84% | motif file (matrix) |
pdf |
| 41 |  | Tbox:Smad/ESCd5-Smad2_3-ChIP-Seq(GSE29422)/Homer | 1e-5 | -1.199e+01 | 0.0000 | 95.0 | 2.99% | 859.5 | 1.85% | motif file (matrix) |
pdf |
| 42 |  | AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-4 | -1.067e+01 | 0.0001 | 236.0 | 7.44% | 2642.0 | 5.68% | motif file (matrix) |
pdf |
| 43 | 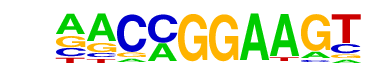 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-4 | -1.045e+01 | 0.0002 | 505.0 | 15.91% | 6237.8 | 13.41% | motif file (matrix) |
pdf |
| 44 |  | Pax7-long(Paired/Homeobox)/Myoblast-Pax7-ChIP-Seq(GSE25064)/Homer | 1e-4 | -1.037e+01 | 0.0002 | 13.0 | 0.41% | 47.2 | 0.10% | motif file (matrix) |
pdf |
| 45 |  | Tcf12(HLH)/GM12878-Tcf12-ChIP-Seq(GSE32465)/Homer | 1e-4 | -9.949e+00 | 0.0003 | 404.0 | 12.73% | 4899.7 | 10.53% | motif file (matrix) |
pdf |
| 46 |  | PAX3:FKHR-fusion(Paired/Homeobox)/Rh4-PAX3:FKHR-ChIP-Seq(GSE19063)/Homer | 1e-4 | -9.645e+00 | 0.0003 | 69.0 | 2.17% | 613.2 | 1.32% | motif file (matrix) |
pdf |
| 47 |  | Oct4:Sox17/F9-Sox17-ChIP-Seq(GSE44553)/Homer | 1e-4 | -9.603e+00 | 0.0004 | 50.0 | 1.58% | 403.3 | 0.87% | motif file (matrix) |
pdf |
| 48 |  | SPDEF(ETS)/VCaP-SPDEF-ChIP-Seq(SRA014231)/Homer | 1e-3 | -8.867e+00 | 0.0007 | 368.0 | 11.59% | 4479.5 | 9.63% | motif file (matrix) |
pdf |
| 49 |  | EBF1(EBF)/Near-E2A-ChIP-Seq(GSE21512)/Homer | 1e-3 | -8.668e+00 | 0.0009 | 401.0 | 12.63% | 4938.0 | 10.61% | motif file (matrix) |
pdf |
| 50 |  | bZIP:IRF/Th17-BatF-ChIP-Seq(GSE39756)/Homer | 1e-3 | -8.554e+00 | 0.0009 | 121.0 | 3.81% | 1264.7 | 2.72% | motif file (matrix) |
pdf |
| 51 |  | Sp1(Zf)/Promoter/Homer | 1e-3 | -8.403e+00 | 0.0011 | 75.0 | 2.36% | 711.9 | 1.53% | motif file (matrix) |
pdf |
| 52 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-3 | -8.340e+00 | 0.0011 | 374.0 | 11.78% | 4593.4 | 9.87% | motif file (matrix) |
pdf |
| 53 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-3 | -8.296e+00 | 0.0012 | 199.0 | 6.27% | 2267.8 | 4.87% | motif file (matrix) |
pdf |
| 54 | 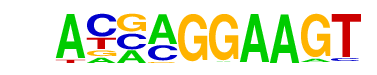 | ELF5(ETS)/T47D-ELF5-ChIP-Seq(GSE30407)/Homer | 1e-3 | -7.896e+00 | 0.0017 | 260.0 | 8.19% | 3089.2 | 6.64% | motif file (matrix) |
pdf |
| 55 |  | Ap4(HLH)/AML-Tfap4-ChIP-Seq(GSE45738)/Homer | 1e-3 | -7.683e+00 | 0.0021 | 472.0 | 14.87% | 5978.4 | 12.85% | motif file (matrix) |
pdf |
| 56 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-3 | -7.374e+00 | 0.0028 | 191.0 | 6.02% | 2206.1 | 4.74% | motif file (matrix) |
pdf |
| 57 |  | Foxh1(Forkhead)/hESC-FOXH1-ChIP-Seq(GSE29422)/Homer | 1e-3 | -7.283e+00 | 0.0030 | 185.0 | 5.83% | 2132.6 | 4.58% | motif file (matrix) |
pdf |
| 58 |  | Rfx2(HTH)/LoVo-RFX2-ChIP-Seq(GSE49402)/Homer | 1e-2 | -6.845e+00 | 0.0045 | 40.0 | 1.26% | 343.1 | 0.74% | motif file (matrix) |
pdf |
| 59 |  | Klf4(Zf)/mES-Klf4-ChIP-Seq(GSE11431)/Homer | 1e-2 | -6.615e+00 | 0.0056 | 144.0 | 4.54% | 1630.6 | 3.50% | motif file (matrix) |
pdf |
| 60 |  | CRE(bZIP)/Promoter/Homer | 1e-2 | -6.540e+00 | 0.0059 | 68.0 | 2.14% | 677.4 | 1.46% | motif file (matrix) |
pdf |
| 61 |  | GATA-DR4(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-2 | -6.225e+00 | 0.0080 | 31.0 | 0.98% | 255.2 | 0.55% | motif file (matrix) |
pdf |
| 62 |  | Rbpj1(?)/Panc1-Rbpj1-ChIP-Seq(GSE47459)/Homer | 1e-2 | -6.030e+00 | 0.0095 | 510.0 | 16.07% | 6642.1 | 14.28% | motif file (matrix) |
pdf |
| 63 |  | EBF(EBF)/proBcell-EBF-ChIP-Seq(GSE21978)/Homer | 1e-2 | -5.989e+00 | 0.0098 | 90.0 | 2.84% | 966.8 | 2.08% | motif file (matrix) |
pdf |
| 64 | 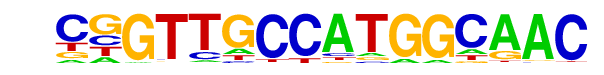 | RFX(HTH)/K562-RFX3-ChIP-Seq(SRA012198)/Homer | 1e-2 | -5.969e+00 | 0.0098 | 35.0 | 1.10% | 304.8 | 0.66% | motif file (matrix) |
pdf |
| 65 |  | Rfx1(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-2 | -5.879e+00 | 0.0106 | 73.0 | 2.30% | 758.5 | 1.63% | motif file (matrix) |
pdf |
| 66 |  | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-2 | -5.736e+00 | 0.0120 | 436.0 | 13.74% | 5638.5 | 12.12% | motif file (matrix) |
pdf |
| 67 |  | CTCF-SatelliteElement/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-2 | -5.578e+00 | 0.0139 | 11.0 | 0.35% | 61.0 | 0.13% | motif file (matrix) |
pdf |
| 68 |  | NeuroD1(bHLH)/Islet-NeuroD1-ChIP-Seq(GSE30298)/Homer | 1e-2 | -5.555e+00 | 0.0140 | 307.0 | 9.67% | 3873.9 | 8.33% | motif file (matrix) |
pdf |
| 69 |  | c-Jun-CRE(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.550e+00 | 0.0140 | 94.0 | 2.96% | 1033.8 | 2.22% | motif file (matrix) |
pdf |
| 70 |  | Nkx3.1(Homeobox)/LNCaP-Nkx3.1-ChIP-Seq(GSE28264)/Homer | 1e-2 | -5.382e+00 | 0.0162 | 759.0 | 23.91% | 10220.8 | 21.97% | motif file (matrix) |
pdf |
| 71 |  | Smad3(MAD)/NPC-Smad3-ChIP-Seq(GSE36673)/Homer | 1e-2 | -5.339e+00 | 0.0166 | 1032.0 | 32.51% | 14131.0 | 30.37% | motif file (matrix) |
pdf |
| 72 | 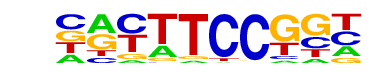 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-2 | -5.159e+00 | 0.0196 | 362.0 | 11.41% | 4663.0 | 10.02% | motif file (matrix) |
pdf |
| 73 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-2 | -5.117e+00 | 0.0202 | 314.0 | 9.89% | 4003.3 | 8.60% | motif file (matrix) |
pdf |
| 74 |  | Bach2(bZIP)/OCILy7-Bach2-ChIP-Seq(GSE44420)/Homer | 1e-2 | -5.097e+00 | 0.0203 | 64.0 | 2.02% | 672.3 | 1.44% | motif file (matrix) |
pdf |
| 75 |  | TCFL2(HMG)/K562-TCF7L2-ChIP-Seq(GSE29196)/Homer | 1e-2 | -5.081e+00 | 0.0204 | 32.0 | 1.01% | 288.2 | 0.62% | motif file (matrix) |
pdf |
| 76 |  | GRHL2(CP2)/HBE-GRHL2-ChIP-Seq(GSE46194)/Homer | 1e-2 | -4.986e+00 | 0.0221 | 116.0 | 3.65% | 1341.9 | 2.88% | motif file (matrix) |
pdf |
| 77 |  | Hoxc9(Homeobox)/Ainv15-Hoxc9-ChIP-Seq(GSE21812)/Homer | 1e-2 | -4.901e+00 | 0.0238 | 131.0 | 4.13% | 1542.9 | 3.32% | motif file (matrix) |
pdf |
| 78 |  | Olig2(bHLH)/Neuron-Olig2-ChIP-Seq(GSE30882)/Homer | 1e-2 | -4.750e+00 | 0.0273 | 663.0 | 20.89% | 8930.8 | 19.20% | motif file (matrix) |
pdf |
| 79 |  | PU.1(ETS)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-2 | -4.628e+00 | 0.0304 | 175.0 | 5.51% | 2145.0 | 4.61% | motif file (matrix) |
pdf |





























































































































































