| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | CTCF(Zf)/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-202 | -4.662e+02 | 0.0000 | 489.0 | 7.99% | 69.8 | 1.42% | motif file (matrix) |
pdf |
| 2 |  | FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-133 | -3.085e+02 | 0.0000 | 1112.0 | 18.16% | 407.9 | 8.27% | motif file (matrix) |
pdf |
| 3 |  | FOXA1(Forkhead)/MCF7-FOXA1-ChIP-Seq(GSE26831)/Homer | 1e-130 | -3.009e+02 | 0.0000 | 936.0 | 15.29% | 316.8 | 6.42% | motif file (matrix) |
pdf |
| 4 |  | BORIS(Zf)/K562-CTCFL-ChIP-Seq(GSE32465)/Homer | 1e-128 | -2.963e+02 | 0.0000 | 514.0 | 8.39% | 117.5 | 2.38% | motif file (matrix) |
pdf |
| 5 |  | Foxa2(Forkhead)/Liver-Foxa2-ChIP-Seq(GSE25694)/Homer | 1e-116 | -2.690e+02 | 0.0000 | 846.0 | 13.82% | 286.1 | 5.80% | motif file (matrix) |
pdf |
| 6 |  | Fox:Ebox(Forkhead:HLH)/Panc1-Foxa2-ChIP-Seq(GSE47459)/Homer | 1e-76 | -1.760e+02 | 0.0000 | 1005.0 | 16.41% | 441.2 | 8.94% | motif file (matrix) |
pdf |
| 7 |  | Sox3(HMG)/NPC-Sox3-ChIP-Seq(GSE33059)/Homer | 1e-65 | -1.511e+02 | 0.0000 | 1467.0 | 23.96% | 765.5 | 15.51% | motif file (matrix) |
pdf |
| 8 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-64 | -1.487e+02 | 0.0000 | 400.0 | 6.53% | 122.4 | 2.48% | motif file (matrix) |
pdf |
| 9 |  | Foxo1(Forkhead)/RAW-Foxo1-ChIP-Seq(Fan et al.)/Homer | 1e-58 | -1.355e+02 | 0.0000 | 1512.0 | 24.69% | 816.7 | 16.55% | motif file (matrix) |
pdf |
| 10 |  | TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer | 1e-54 | -1.255e+02 | 0.0000 | 764.0 | 12.48% | 340.7 | 6.90% | motif file (matrix) |
pdf |
| 11 |  | TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer | 1e-53 | -1.224e+02 | 0.0000 | 587.0 | 9.59% | 238.4 | 4.83% | motif file (matrix) |
pdf |
| 12 |  | Sox2(HMG)/mES-Sox2-ChIP-Seq(GSE11431)/Homer | 1e-48 | -1.116e+02 | 0.0000 | 794.0 | 12.97% | 372.8 | 7.56% | motif file (matrix) |
pdf |
| 13 |  | Sox6(HMG)/Myotubes-Sox6-ChIP-Seq(GSE32627)/Homer | 1e-47 | -1.101e+02 | 0.0000 | 1253.0 | 20.46% | 674.8 | 13.68% | motif file (matrix) |
pdf |
| 14 |  | Gata2(Zf)/K562-GATA2-ChIP-Seq(GSE18829)/Homer | 1e-47 | -1.088e+02 | 0.0000 | 551.0 | 9.00% | 228.4 | 4.63% | motif file (matrix) |
pdf |
| 15 |  | Gata4(Zf)/Heart-Gata4-ChIP-Seq(GSE35151)/Homer | 1e-47 | -1.082e+02 | 0.0000 | 737.0 | 12.04% | 340.7 | 6.91% | motif file (matrix) |
pdf |
| 16 |  | Gata1(Zf)/K562-GATA1-ChIP-Seq(GSE18829)/Homer | 1e-42 | -9.746e+01 | 0.0000 | 504.0 | 8.23% | 210.1 | 4.26% | motif file (matrix) |
pdf |
| 17 |  | GATA3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-42 | -9.692e+01 | 0.0000 | 997.0 | 16.28% | 520.8 | 10.55% | motif file (matrix) |
pdf |
| 18 |  | Tcf3(HMG)/mES-Tcf3-ChIP-Seq(GSE11724)/Homer | 1e-30 | -6.942e+01 | 0.0000 | 223.0 | 3.64% | 75.9 | 1.54% | motif file (matrix) |
pdf |
| 19 |  | Rfx2(HTH)/LoVo-RFX2-ChIP-Seq(GSE49402)/Homer | 1e-23 | -5.335e+01 | 0.0000 | 133.0 | 2.17% | 39.3 | 0.80% | motif file (matrix) |
pdf |
| 20 |  | Hnf1(Homeobox)/Liver-Foxa2-Chip-Seq(GSE25694)/Homer | 1e-21 | -4.901e+01 | 0.0000 | 91.0 | 1.49% | 22.1 | 0.45% | motif file (matrix) |
pdf |
| 21 |  | X-box(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-20 | -4.805e+01 | 0.0000 | 136.0 | 2.22% | 43.6 | 0.88% | motif file (matrix) |
pdf |
| 22 | 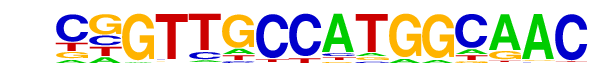 | RFX(HTH)/K562-RFX3-ChIP-Seq(SRA012198)/Homer | 1e-19 | -4.442e+01 | 0.0000 | 122.0 | 1.99% | 38.1 | 0.77% | motif file (matrix) |
pdf |
| 23 |  | Lhx2(Homeobox)/HFSC-Lhx2-ChIP-Seq(GSE48068)/Homer | 1e-18 | -4.335e+01 | 0.0000 | 574.0 | 9.37% | 315.9 | 6.40% | motif file (matrix) |
pdf |
| 24 |  | Klf4(Zf)/mES-Klf4-ChIP-Seq(GSE11431)/Homer | 1e-18 | -4.242e+01 | 0.0000 | 335.0 | 5.47% | 161.9 | 3.28% | motif file (matrix) |
pdf |
| 25 |  | Lhx3(Homeobox)/Neuron-Lhx3-ChIP-Seq(GSE31456)/Homer | 1e-17 | -4.080e+01 | 0.0000 | 885.0 | 14.45% | 535.1 | 10.85% | motif file (matrix) |
pdf |
| 26 |  | Rfx5(HTH)/GM12878-Rfx5-ChIP-Seq(GSE31477)/Homer | 1e-17 | -3.921e+01 | 0.0000 | 279.0 | 4.56% | 130.2 | 2.64% | motif file (matrix) |
pdf |
| 27 |  | TCFL2(HMG)/K562-TCF7L2-ChIP-Seq(GSE29196)/Homer | 1e-16 | -3.865e+01 | 0.0000 | 91.0 | 1.49% | 26.3 | 0.53% | motif file (matrix) |
pdf |
| 28 |  | KLF5(Zf)/LoVo-KLF5-ChIP-Seq(GSE49402)/Homer | 1e-15 | -3.609e+01 | 0.0000 | 1041.0 | 17.00% | 658.6 | 13.35% | motif file (matrix) |
pdf |
| 29 |  | CTCF-SatelliteElement/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-14 | -3.269e+01 | 0.0000 | 27.0 | 0.44% | 3.8 | 0.08% | motif file (matrix) |
pdf |
| 30 |  | Unknown-ESC-element/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-14 | -3.256e+01 | 0.0000 | 567.0 | 9.26% | 329.4 | 6.68% | motif file (matrix) |
pdf |
| 31 |  | Tcf4(HMG)/Hct116-Tcf4-ChIP-Seq(SRA012054)/Homer | 1e-13 | -3.155e+01 | 0.0000 | 366.0 | 5.98% | 195.8 | 3.97% | motif file (matrix) |
pdf |
| 32 |  | Nkx6.1(Homeobox)/Islet-Nkx6.1-ChIP-Seq(GSE40975)/Homer | 1e-13 | -3.119e+01 | 0.0000 | 1310.0 | 21.39% | 870.3 | 17.64% | motif file (matrix) |
pdf |
| 33 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-12 | -2.938e+01 | 0.0000 | 358.0 | 5.85% | 193.6 | 3.92% | motif file (matrix) |
pdf |
| 34 |  | REST-NRSF(Zf)/Jurkat-NRSF-ChIP-Seq/Homer | 1e-12 | -2.932e+01 | 0.0000 | 38.0 | 0.62% | 7.8 | 0.16% | motif file (matrix) |
pdf |
| 35 |  | Sp1(Zf)/Promoter/Homer | 1e-12 | -2.887e+01 | 0.0000 | 197.0 | 3.22% | 91.4 | 1.85% | motif file (matrix) |
pdf |
| 36 |  | c-Jun-CRE(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-11 | -2.744e+01 | 0.0000 | 183.0 | 2.99% | 84.5 | 1.71% | motif file (matrix) |
pdf |
| 37 |  | Rfx1(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-11 | -2.686e+01 | 0.0000 | 200.0 | 3.27% | 95.3 | 1.93% | motif file (matrix) |
pdf |
| 38 |  | EKLF(Zf)/Erythrocyte-Klf1-ChIP-Seq(GSE20478)/Homer | 1e-10 | -2.445e+01 | 0.0000 | 181.0 | 2.96% | 86.8 | 1.76% | motif file (matrix) |
pdf |
| 39 |  | AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-10 | -2.444e+01 | 0.0000 | 402.0 | 6.57% | 231.8 | 4.70% | motif file (matrix) |
pdf |
| 40 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-10 | -2.430e+01 | 0.0000 | 356.0 | 5.81% | 200.2 | 4.06% | motif file (matrix) |
pdf |
| 41 |  | FXR(NR/IR1)/Liver-FXR-ChIP-Seq(Chong et al.)/Homer | 1e-10 | -2.315e+01 | 0.0000 | 257.0 | 4.20% | 136.0 | 2.76% | motif file (matrix) |
pdf |
| 42 |  | MafA(bZIP)/Islet-MafA-ChIP-Seq(GSE30298)/Homer | 1e-9 | -2.282e+01 | 0.0000 | 573.0 | 9.36% | 354.1 | 7.18% | motif file (matrix) |
pdf |
| 43 |  | Bach2(bZIP)/OCILy7-Bach2-ChIP-Seq(GSE44420)/Homer | 1e-9 | -2.249e+01 | 0.0000 | 133.0 | 2.17% | 59.2 | 1.20% | motif file (matrix) |
pdf |
| 44 | 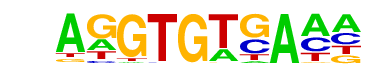 | Tbet(T-box)/CD8-Tbet-ChIP-Seq(GSE33802)/Homer | 1e-9 | -2.101e+01 | 0.0000 | 646.0 | 10.55% | 411.8 | 8.35% | motif file (matrix) |
pdf |
| 45 |  | Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-8 | -2.029e+01 | 0.0000 | 136.0 | 2.22% | 63.9 | 1.29% | motif file (matrix) |
pdf |
| 46 |  | Oct2(POU/Homeobox)/Bcell-Oct2-ChIP-Seq(GSE21512)/Homer | 1e-8 | -1.875e+01 | 0.0000 | 130.0 | 2.12% | 61.6 | 1.25% | motif file (matrix) |
pdf |
| 47 |  | Atf1(bZIP)/K562-ATF1-ChIP-Seq(GSE31477)/Homer | 1e-8 | -1.854e+01 | 0.0000 | 369.0 | 6.03% | 220.8 | 4.47% | motif file (matrix) |
pdf |
| 48 |  | Tbx20(T-box)/Heart-Tbx20-ChIP-Seq(GSE29636)/Homer | 1e-7 | -1.661e+01 | 0.0000 | 184.0 | 3.01% | 98.3 | 1.99% | motif file (matrix) |
pdf |
| 49 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-6 | -1.569e+01 | 0.0000 | 330.0 | 5.39% | 199.3 | 4.04% | motif file (matrix) |
pdf |
| 50 |  | Maz(Zf)/HepG2-Maz-ChIP-Seq(GSE31477)/Homer | 1e-6 | -1.555e+01 | 0.0000 | 1177.0 | 19.22% | 826.4 | 16.75% | motif file (matrix) |
pdf |
| 51 |  | Oct4:Sox17/F9-Sox17-ChIP-Seq(GSE44553)/Homer | 1e-6 | -1.472e+01 | 0.0000 | 85.0 | 1.39% | 38.8 | 0.79% | motif file (matrix) |
pdf |
| 52 |  | NFY(CCAAT)/Promoter/Homer | 1e-6 | -1.444e+01 | 0.0000 | 434.0 | 7.09% | 276.6 | 5.61% | motif file (matrix) |
pdf |
| 53 |  | CRX(Homeobox)/Retina-Crx-ChIP-Seq(GSE20012)/Homer | 1e-6 | -1.396e+01 | 0.0000 | 1285.0 | 20.99% | 916.4 | 18.57% | motif file (matrix) |
pdf |
| 54 |  | Znf263(Zf)/K562-Znf263-ChIP-Seq(GSE31477)/Homer | 1e-5 | -1.360e+01 | 0.0000 | 1358.0 | 22.18% | 974.7 | 19.75% | motif file (matrix) |
pdf |
| 55 |  | JunD(bZIP)/K562-JunD-ChIP-Seq/Homer | 1e-5 | -1.347e+01 | 0.0000 | 59.0 | 0.96% | 24.7 | 0.50% | motif file (matrix) |
pdf |
| 56 |  | Nanog(Homeobox)/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-5 | -1.307e+01 | 0.0000 | 2416.0 | 39.46% | 1806.5 | 36.61% | motif file (matrix) |
pdf |
| 57 |  | MafK(bZIP)/C2C12-MafK-ChIP-Seq(GSE36030)/Homer | 1e-5 | -1.277e+01 | 0.0000 | 137.0 | 2.24% | 73.3 | 1.48% | motif file (matrix) |
pdf |
| 58 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-5 | -1.242e+01 | 0.0000 | 333.0 | 5.44% | 209.8 | 4.25% | motif file (matrix) |
pdf |
| 59 |  | Eomes(T-box)/H9-Eomes-ChIP-Seq(GSE26097)/Homer | 1e-5 | -1.210e+01 | 0.0000 | 1278.0 | 20.87% | 920.3 | 18.65% | motif file (matrix) |
pdf |
| 60 |  | GATA:SCL/Ter119-SCL-ChIP-Seq(GSE18720)/Homer | 1e-5 | -1.200e+01 | 0.0000 | 82.0 | 1.34% | 39.7 | 0.80% | motif file (matrix) |
pdf |
| 61 |  | Erra(NR)/HepG2-Erra-ChIP-Seq(GSE31477)/Homer | 1e-4 | -1.147e+01 | 0.0000 | 1480.0 | 24.17% | 1080.7 | 21.90% | motif file (matrix) |
pdf |
| 62 |  | ZBTB33(Zf)/GM12878-ZBTB33-ChIP-Seq(GSE32465)/Homer | 1e-4 | -1.093e+01 | 0.0001 | 17.0 | 0.28% | 4.4 | 0.09% | motif file (matrix) |
pdf |
| 63 |  | NRF1/Promoter/Homer | 1e-4 | -1.091e+01 | 0.0001 | 80.0 | 1.31% | 39.9 | 0.81% | motif file (matrix) |
pdf |
| 64 |  | NeuroD1(bHLH)/Islet-NeuroD1-ChIP-Seq(GSE30298)/Homer | 1e-4 | -1.065e+01 | 0.0001 | 614.0 | 10.03% | 421.2 | 8.54% | motif file (matrix) |
pdf |
| 65 |  | NFkB-p65-Rel(RHD)/LPS-exp(GSE23622)/Homer | 1e-4 | -1.003e+01 | 0.0002 | 40.0 | 0.65% | 16.7 | 0.34% | motif file (matrix) |
pdf |
| 66 |  | Pitx1(Homeobox)/Chicken-Pitx1-ChIP-Seq(GSE38910)/Homer | 1e-4 | -9.853e+00 | 0.0002 | 2347.0 | 38.33% | 1773.2 | 35.94% | motif file (matrix) |
pdf |
| 67 |  | Isl1(Homeobox)/Neuron-Isl1-ChIP-Seq(GSE31456)/Homer | 1e-4 | -9.789e+00 | 0.0002 | 1114.0 | 18.19% | 806.0 | 16.34% | motif file (matrix) |
pdf |
| 68 |  | Atf4(bZIP)/MEF-Atf4-ChIP-Seq(GSE35681)/Homer | 1e-4 | -9.671e+00 | 0.0002 | 119.0 | 1.94% | 66.3 | 1.34% | motif file (matrix) |
pdf |
| 69 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-4 | -9.455e+00 | 0.0003 | 675.0 | 11.02% | 472.4 | 9.57% | motif file (matrix) |
pdf |
| 70 |  | Nrf2(bZIP)/Lymphoblast-Nrf2-ChIP-Seq(GSE37589)/Homer | 1e-3 | -9.189e+00 | 0.0004 | 26.0 | 0.42% | 9.6 | 0.19% | motif file (matrix) |
pdf |
| 71 |  | Olig2(bHLH)/Neuron-Olig2-ChIP-Seq(GSE30882)/Homer | 1e-3 | -9.121e+00 | 0.0004 | 1251.0 | 20.43% | 916.5 | 18.58% | motif file (matrix) |
pdf |
| 72 |  | GRHL2(CP2)/HBE-GRHL2-ChIP-Seq(GSE46194)/Homer | 1e-3 | -9.115e+00 | 0.0004 | 248.0 | 4.05% | 157.4 | 3.19% | motif file (matrix) |
pdf |
| 73 |  | Rbpj1(?)/Panc1-Rbpj1-ChIP-Seq(GSE47459)/Homer | 1e-3 | -8.831e+00 | 0.0005 | 963.0 | 15.73% | 695.4 | 14.09% | motif file (matrix) |
pdf |
| 74 |  | Six1(Homeobox)/Myoblast-Six1-ChIP-Chip(GSE20150)/Homer | 1e-3 | -8.798e+00 | 0.0005 | 149.0 | 2.43% | 88.0 | 1.78% | motif file (matrix) |
pdf |
| 75 |  | Nur77(NR)/K562-NR4A1-ChIP-Seq(GSE31363)/Homer | 1e-3 | -8.739e+00 | 0.0005 | 124.0 | 2.03% | 71.6 | 1.45% | motif file (matrix) |
pdf |
| 76 |  | PRDM14(Zf)/H1-PRDM14-ChIP-Seq(GSE22767)/Homer | 1e-3 | -8.521e+00 | 0.0006 | 215.0 | 3.51% | 135.2 | 2.74% | motif file (matrix) |
pdf |
| 77 |  | Tbx5(T-box)/HL1-Tbx5.biotin-ChIP-Seq(GSE21529)/Homer | 1e-3 | -8.506e+00 | 0.0006 | 2086.0 | 34.07% | 1576.2 | 31.95% | motif file (matrix) |
pdf |
| 78 |  | SCL(HLH)/HPC7-Scl-ChIP-Seq(GSE13511)/Homer | 1e-3 | -8.491e+00 | 0.0006 | 3127.0 | 51.07% | 2408.2 | 48.81% | motif file (matrix) |
pdf |
| 79 |  | MafF(bZIP)/HepG2-MafF-ChIP-Seq(GSE31477)/Homer | 1e-3 | -8.471e+00 | 0.0007 | 107.0 | 1.75% | 60.7 | 1.23% | motif file (matrix) |
pdf |
| 80 |  | HNF6(Homeobox)/Liver-Hnf6-ChIP-Seq(ERP000394)/Homer | 1e-3 | -8.412e+00 | 0.0007 | 223.0 | 3.64% | 141.3 | 2.86% | motif file (matrix) |
pdf |
| 81 |  | RUNX(Runt)/HPC7-Runx1-ChIP-Seq(GSE22178)/Homer | 1e-3 | -8.377e+00 | 0.0007 | 456.0 | 7.45% | 312.7 | 6.34% | motif file (matrix) |
pdf |
| 82 |  | ETS(ETS)/Promoter/Homer | 1e-3 | -8.270e+00 | 0.0008 | 193.0 | 3.15% | 120.8 | 2.45% | motif file (matrix) |
pdf |
| 83 |  | Oct4(POU/Homeobox)/mES-Oct4-ChIP-Seq(GSE11431)/Homer | 1e-3 | -8.155e+00 | 0.0009 | 194.0 | 3.17% | 121.8 | 2.47% | motif file (matrix) |
pdf |
| 84 |  | Smad2(MAD)/ES-SMAD2-ChIP-Seq(GSE29422)/Homer | 1e-3 | -7.964e+00 | 0.0010 | 1029.0 | 16.81% | 751.8 | 15.24% | motif file (matrix) |
pdf |
| 85 | 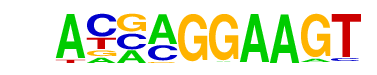 | ELF5(ETS)/T47D-ELF5-ChIP-Seq(GSE30407)/Homer | 1e-3 | -7.887e+00 | 0.0011 | 448.0 | 7.32% | 308.7 | 6.26% | motif file (matrix) |
pdf |
| 86 |  | BMYB(HTH)/Hela-BMYB-ChIPSeq(GSE27030)/Homer | 1e-3 | -7.876e+00 | 0.0011 | 891.0 | 14.55% | 645.3 | 13.08% | motif file (matrix) |
pdf |
| 87 |  | E2A-nearPU.1(HLH)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-3 | -7.722e+00 | 0.0013 | 943.0 | 15.40% | 686.1 | 13.90% | motif file (matrix) |
pdf |
| 88 |  | GATA-IR3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-3 | -7.474e+00 | 0.0016 | 76.0 | 1.24% | 42.0 | 0.85% | motif file (matrix) |
pdf |
| 89 |  | Ptf1a(HLH)/Panc1-Ptf1a-ChIP-Seq(GSE47459)/Homer | 1e-3 | -7.332e+00 | 0.0018 | 1838.0 | 30.02% | 1389.1 | 28.15% | motif file (matrix) |
pdf |
| 90 |  | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-3 | -7.320e+00 | 0.0018 | 901.0 | 14.72% | 656.2 | 13.30% | motif file (matrix) |
pdf |
| 91 |  | GFY-Staf/Promoters/Homer | 1e-3 | -7.241e+00 | 0.0019 | 16.0 | 0.26% | 5.4 | 0.11% | motif file (matrix) |
pdf |
| 92 |  | CArG(MADS)/PUER-Srf-ChIP-Seq(Sullivan et al.)/Homer | 1e-3 | -7.197e+00 | 0.0020 | 144.0 | 2.35% | 88.1 | 1.79% | motif file (matrix) |
pdf |
| 93 |  | PBX1(Homeobox)/MCF7-PBX1-ChIP-Seq(GSE28007)/Homer | 1e-3 | -7.187e+00 | 0.0020 | 60.0 | 0.98% | 31.3 | 0.63% | motif file (matrix) |
pdf |
| 94 |  | RUNX2(Runt)/PCa-RUNX2-ChIP-Seq(GSE33889)/Homer | 1e-3 | -6.998e+00 | 0.0024 | 500.0 | 8.17% | 351.3 | 7.12% | motif file (matrix) |
pdf |
| 95 |  | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-2 | -6.762e+00 | 0.0030 | 789.0 | 12.89% | 574.0 | 11.63% | motif file (matrix) |
pdf |
| 96 |  | E2F6(E2F)/Hela-E2F6-ChIP-Seq(GSE31477)/Homer | 1e-2 | -6.712e+00 | 0.0031 | 294.0 | 4.80% | 198.6 | 4.02% | motif file (matrix) |
pdf |
| 97 |  | HOXA2(Homeobox)/mES-Hoxa2-ChIP-Seq(Donaldson et al.)/Homer | 1e-2 | -6.463e+00 | 0.0040 | 46.0 | 0.75% | 23.1 | 0.47% | motif file (matrix) |
pdf |
| 98 |  | Pax7-long(Paired/Homeobox)/Myoblast-Pax7-ChIP-Seq(GSE25064)/Homer | 1e-2 | -6.412e+00 | 0.0041 | 11.0 | 0.18% | 3.7 | 0.07% | motif file (matrix) |
pdf |
| 99 |  | E2A(HLH)/proBcell-E2A-ChIP-Seq(GSE21978)/Homer | 1e-2 | -6.343e+00 | 0.0044 | 1077.0 | 17.59% | 799.4 | 16.20% | motif file (matrix) |
pdf |
| 100 |  | Chop(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer | 1e-2 | -6.319e+00 | 0.0044 | 88.0 | 1.44% | 51.5 | 1.04% | motif file (matrix) |
pdf |
| 101 |  | PAX5(Paired/Homeobox)/GM12878-PAX5-ChIP-Seq(GSE32465)/Homer | 1e-2 | -6.299e+00 | 0.0045 | 209.0 | 3.41% | 137.1 | 2.78% | motif file (matrix) |
pdf |
| 102 |  | NF1:FOXA1/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-2 | -6.260e+00 | 0.0046 | 36.0 | 0.59% | 17.6 | 0.36% | motif file (matrix) |
pdf |
| 103 |  | GATA-IR4(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-2 | -6.246e+00 | 0.0046 | 44.0 | 0.72% | 22.5 | 0.46% | motif file (matrix) |
pdf |
| 104 |  | ETS:E-box/HPC7-Scl-ChIP-Seq(GSE22178)/Homer | 1e-2 | -5.807e+00 | 0.0071 | 63.0 | 1.03% | 35.1 | 0.71% | motif file (matrix) |
pdf |
| 105 |  | CEBP:AP1(bZIP)/ThioMac-CEBPb-ChIP-Seq(GSE21512)/Homer | 1e-2 | -5.770e+00 | 0.0073 | 336.0 | 5.49% | 233.4 | 4.73% | motif file (matrix) |
pdf |
| 106 |  | CRE(bZIP)/Promoter/Homer | 1e-2 | -5.718e+00 | 0.0076 | 122.0 | 1.99% | 76.4 | 1.55% | motif file (matrix) |
pdf |
| 107 |  | RUNX-AML(Runt)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer | 1e-2 | -5.671e+00 | 0.0079 | 444.0 | 7.25% | 315.7 | 6.40% | motif file (matrix) |
pdf |
| 108 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-2 | -5.618e+00 | 0.0083 | 589.0 | 9.62% | 426.1 | 8.63% | motif file (matrix) |
pdf |
| 109 |  | Tcf12(HLH)/GM12878-Tcf12-ChIP-Seq(GSE32465)/Homer | 1e-2 | -5.335e+00 | 0.0109 | 763.0 | 12.46% | 562.0 | 11.39% | motif file (matrix) |
pdf |
| 110 |  | p53(p53)/mES-cMyc-ChIP-Seq(GSE11431)/Homer | 1e-2 | -5.290e+00 | 0.0113 | 10.0 | 0.16% | 3.2 | 0.07% | motif file (matrix) |
pdf |
| 111 |  | p53(p53)/Saos-p53-ChIP-Seq(GSE15780)/Homer | 1e-2 | -5.192e+00 | 0.0122 | 48.0 | 0.78% | 26.5 | 0.54% | motif file (matrix) |
pdf |
| 112 |  | p53(p53)/Saos-p53-ChIP-Seq/Homer | 1e-2 | -5.192e+00 | 0.0122 | 48.0 | 0.78% | 26.5 | 0.54% | motif file (matrix) |
pdf |
| 113 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.148e+00 | 0.0126 | 331.0 | 5.41% | 232.5 | 4.71% | motif file (matrix) |
pdf |
| 114 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-2 | -5.127e+00 | 0.0128 | 1069.0 | 17.46% | 802.5 | 16.26% | motif file (matrix) |
pdf |
| 115 | 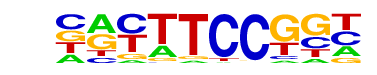 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-2 | -5.072e+00 | 0.0134 | 731.0 | 11.94% | 539.7 | 10.94% | motif file (matrix) |
pdf |
| 116 |  | BMAL1(HLH)/Liver-Bmal1-ChIP-Seq(GSE39860)/Homer | 1e-2 | -5.054e+00 | 0.0135 | 916.0 | 14.96% | 683.7 | 13.86% | motif file (matrix) |
pdf |
| 117 |  | ZFX(Zf)/mES-Zfx-ChIP-Seq(GSE11431)/Homer | 1e-2 | -4.903e+00 | 0.0156 | 894.0 | 14.60% | 667.0 | 13.52% | motif file (matrix) |
pdf |
| 118 |  | Smad4(MAD)/ESC-SMAD4-ChIP-Seq(GSE29422)/Homer | 1e-2 | -4.725e+00 | 0.0185 | 1066.0 | 17.41% | 803.6 | 16.29% | motif file (matrix) |
pdf |











































































































































































































































