| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | E2F7(E2F)/Hela-E2F7-ChIP-Seq(GSE32673)/Homer | 1e-10 | -2.385e+01 | 0.0000 | 26.0 | 1.04% | 2.6 | 0.27% | motif file (matrix) |
pdf |
| 2 |  | FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-9 | -2.173e+01 | 0.0000 | 1187.0 | 47.54% | 405.5 | 41.46% | motif file (matrix) |
pdf |
| 3 |  | Foxa2(Forkhead)/Liver-Foxa2-ChIP-Seq(GSE25694)/Homer | 1e-9 | -2.132e+01 | 0.0000 | 739.0 | 29.60% | 237.9 | 24.32% | motif file (matrix) |
pdf |
| 4 |  | RUNX1(Runt)/Jurkat-RUNX1-ChIP-Seq(GSE29180)/Homer | 1e-7 | -1.724e+01 | 0.0000 | 691.0 | 27.67% | 225.6 | 23.06% | motif file (matrix) |
pdf |
| 5 |  | Olig2(bHLH)/Neuron-Olig2-ChIP-Seq(GSE30882)/Homer | 1e-7 | -1.651e+01 | 0.0000 | 1196.0 | 47.90% | 417.9 | 42.73% | motif file (matrix) |
pdf |
| 6 |  | FOXA1(Forkhead)/MCF7-FOXA1-ChIP-Seq(GSE26831)/Homer | 1e-6 | -1.409e+01 | 0.0000 | 1039.0 | 41.61% | 361.1 | 36.92% | motif file (matrix) |
pdf |
| 7 |  | Nkx2.1(Homeobox)/LungAC-Nkx2.1-ChIP-Seq(GSE43252)/Homer | 1e-5 | -1.299e+01 | 0.0001 | 1668.0 | 66.80% | 610.1 | 62.38% | motif file (matrix) |
pdf |
| 8 |  | CEBP(bZIP)/CEBPb-ChIP-Seq(GSE21512)/Homer | 1e-5 | -1.265e+01 | 0.0001 | 752.0 | 30.12% | 255.6 | 26.13% | motif file (matrix) |
pdf |
| 9 |  | HIF2a(HLH)/785_O-HIF2a-ChIP-Seq(GSE34871)/Homer | 1e-5 | -1.167e+01 | 0.0002 | 172.0 | 6.89% | 48.6 | 4.96% | motif file (matrix) |
pdf |
| 10 |  | Esrrb(NR)/mES-Esrrb-ChIP-Seq(GSE11431)/Homer | 1e-4 | -1.101e+01 | 0.0004 | 438.0 | 17.54% | 142.8 | 14.60% | motif file (matrix) |
pdf |
| 11 |  | GRHL2(CP2)/HBE-GRHL2-ChIP-Seq(GSE46194)/Homer | 1e-4 | -1.095e+01 | 0.0004 | 295.0 | 11.81% | 91.9 | 9.39% | motif file (matrix) |
pdf |
| 12 |  | AP-2gamma(AP2)/MCF7-TFAP2C-ChIP-Seq(GSE21234)/Homer | 1e-4 | -1.083e+01 | 0.0004 | 404.0 | 16.18% | 130.7 | 13.36% | motif file (matrix) |
pdf |
| 13 |  | Nkx2.5(Homeobox)/HL1-Nkx2.5.biotin-ChIP-Seq(GSE21529)/Homer | 1e-4 | -1.081e+01 | 0.0004 | 1448.0 | 57.99% | 527.3 | 53.91% | motif file (matrix) |
pdf |
| 14 |  | EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer | 1e-4 | -1.068e+01 | 0.0004 | 565.0 | 22.63% | 189.2 | 19.35% | motif file (matrix) |
pdf |
| 15 | 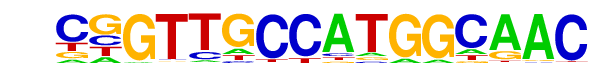 | RFX(HTH)/K562-RFX3-ChIP-Seq(SRA012198)/Homer | 1e-4 | -1.056e+01 | 0.0004 | 30.0 | 1.20% | 5.9 | 0.61% | motif file (matrix) |
pdf |
| 16 |  | E2F4(E2F)/K562-E2F4-ChIP-Seq(GSE31477)/Homer | 1e-4 | -1.023e+01 | 0.0006 | 101.0 | 4.04% | 26.1 | 2.67% | motif file (matrix) |
pdf |
| 17 |  | PAX5(Paired/Homeobox)/GM12878-PAX5-ChIP-Seq(GSE32465)/Homer | 1e-4 | -1.006e+01 | 0.0006 | 197.0 | 7.89% | 58.7 | 6.00% | motif file (matrix) |
pdf |
| 18 |  | MyoD(HLH)/Myotube-MyoD-ChIP-Seq(GSE21614)/Homer | 1e-4 | -9.975e+00 | 0.0006 | 314.0 | 12.58% | 99.2 | 10.14% | motif file (matrix) |
pdf |
| 19 |  | VDR(NR/DR3)/GM10855-VDR+vitD-ChIP-Seq(GSE22484)/Homer | 1e-4 | -9.860e+00 | 0.0007 | 131.0 | 5.25% | 36.3 | 3.71% | motif file (matrix) |
pdf |
| 20 |  | NRF1/Promoter/Homer | 1e-4 | -9.680e+00 | 0.0008 | 29.0 | 1.16% | 5.8 | 0.60% | motif file (matrix) |
pdf |
| 21 |  | RUNX(Runt)/HPC7-Runx1-ChIP-Seq(GSE22178)/Homer | 1e-4 | -9.597e+00 | 0.0008 | 440.0 | 17.62% | 145.2 | 14.85% | motif file (matrix) |
pdf |
| 22 |  | Fox:Ebox(Forkhead:HLH)/Panc1-Foxa2-ChIP-Seq(GSE47459)/Homer | 1e-4 | -9.550e+00 | 0.0008 | 744.0 | 29.80% | 258.8 | 26.46% | motif file (matrix) |
pdf |
| 23 |  | Mouse_Recombination_Hotspot/Testis-DMC1-ChIP-Seq(GSE24438)/Homer | 1e-3 | -9.136e+00 | 0.0012 | 70.0 | 2.80% | 17.5 | 1.79% | motif file (matrix) |
pdf |
| 24 |  | ZNF711(Zf)/SH-SY5Y-ZNF711-ChIP-Seq(GSE20673)/Homer | 1e-3 | -9.042e+00 | 0.0012 | 794.0 | 31.80% | 279.0 | 28.52% | motif file (matrix) |
pdf |
| 25 |  | CEBP:AP1(bZIP)/ThioMac-CEBPb-ChIP-Seq(GSE21512)/Homer | 1e-3 | -8.980e+00 | 0.0012 | 685.0 | 27.43% | 237.3 | 24.27% | motif file (matrix) |
pdf |
| 26 |  | Atf1(bZIP)/K562-ATF1-ChIP-Seq(GSE31477)/Homer | 1e-3 | -8.905e+00 | 0.0013 | 583.0 | 23.35% | 199.1 | 20.36% | motif file (matrix) |
pdf |
| 27 |  | RARg(NR)/ES-RARg-ChIP-Seq(GSE30538)/Homer | 1e-3 | -8.865e+00 | 0.0013 | 20.0 | 0.80% | 3.6 | 0.37% | motif file (matrix) |
pdf |
| 28 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-3 | -8.723e+00 | 0.0014 | 184.0 | 7.37% | 55.7 | 5.69% | motif file (matrix) |
pdf |
| 29 |  | Foxo1(Forkhead)/RAW-Foxo1-ChIP-Seq(Fan et al.)/Homer | 1e-3 | -8.600e+00 | 0.0016 | 1228.0 | 49.18% | 446.4 | 45.64% | motif file (matrix) |
pdf |
| 30 |  | ZFX(Zf)/mES-Zfx-ChIP-Seq(GSE11431)/Homer | 1e-3 | -8.528e+00 | 0.0016 | 589.0 | 23.59% | 202.1 | 20.66% | motif file (matrix) |
pdf |
| 31 |  | ETS(ETS)/Promoter/Homer | 1e-3 | -8.366e+00 | 0.0018 | 130.0 | 5.21% | 37.2 | 3.80% | motif file (matrix) |
pdf |
| 32 |  | HRE(HSF)/HepG2-HSF1-ChIP-Seq(GSE31477)/Homer | 1e-3 | -8.180e+00 | 0.0022 | 87.0 | 3.48% | 23.1 | 2.36% | motif file (matrix) |
pdf |
| 33 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-3 | -8.019e+00 | 0.0025 | 446.0 | 17.86% | 150.5 | 15.39% | motif file (matrix) |
pdf |
| 34 |  | Tcf3(HMG)/mES-Tcf3-ChIP-Seq(GSE11724)/Homer | 1e-3 | -7.927e+00 | 0.0026 | 210.0 | 8.41% | 65.6 | 6.71% | motif file (matrix) |
pdf |
| 35 |  | ETS:RUNX/Jurkat-RUNX1-ChIP-Seq(GSE17954)/Homer | 1e-3 | -7.786e+00 | 0.0029 | 34.0 | 1.36% | 7.8 | 0.79% | motif file (matrix) |
pdf |
| 36 |  | PR(NR)/T47D-PR-ChIP-Seq(GSE31130)/Homer | 1e-3 | -7.491e+00 | 0.0038 | 1264.0 | 50.62% | 463.6 | 47.40% | motif file (matrix) |
pdf |
| 37 |  | Oct4:Sox17/F9-Sox17-ChIP-Seq(GSE44553)/Homer | 1e-3 | -7.435e+00 | 0.0039 | 171.0 | 6.85% | 52.1 | 5.33% | motif file (matrix) |
pdf |
| 38 |  | bZIP:IRF/Th17-BatF-ChIP-Seq(GSE39756)/Homer | 1e-3 | -7.346e+00 | 0.0042 | 483.0 | 19.34% | 165.1 | 16.89% | motif file (matrix) |
pdf |
| 39 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.225e+00 | 0.0046 | 196.0 | 7.85% | 61.1 | 6.24% | motif file (matrix) |
pdf |
| 40 |  | Smad3(MAD)/NPC-Smad3-ChIP-Seq(GSE36673)/Homer | 1e-3 | -7.158e+00 | 0.0048 | 1412.0 | 56.55% | 522.9 | 53.47% | motif file (matrix) |
pdf |
| 41 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-3 | -7.034e+00 | 0.0053 | 567.0 | 22.71% | 197.9 | 20.23% | motif file (matrix) |
pdf |
| 42 |  | Sox2(HMG)/mES-Sox2-ChIP-Seq(GSE11431)/Homer | 1e-2 | -6.851e+00 | 0.0062 | 717.0 | 28.71% | 254.8 | 26.05% | motif file (matrix) |
pdf |
| 43 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-2 | -6.736e+00 | 0.0068 | 458.0 | 18.34% | 157.5 | 16.10% | motif file (matrix) |
pdf |
| 44 |  | CTCF-SatelliteElement/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-2 | -6.641e+00 | 0.0073 | 9.0 | 0.36% | 0.9 | 0.10% | motif file (matrix) |
pdf |
| 45 |  | RUNX-AML(Runt)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer | 1e-2 | -6.593e+00 | 0.0075 | 476.0 | 19.06% | 164.3 | 16.80% | motif file (matrix) |
pdf |
| 46 |  | MafK(bZIP)/C2C12-MafK-ChIP-Seq(GSE36030)/Homer | 1e-2 | -6.557e+00 | 0.0076 | 104.0 | 4.16% | 31.0 | 3.17% | motif file (matrix) |
pdf |
| 47 |  | USF1(HLH)/GM12878-Usf1-ChIP-Seq(GSE32465)/Homer | 1e-2 | -6.201e+00 | 0.0106 | 192.0 | 7.69% | 61.5 | 6.29% | motif file (matrix) |
pdf |
| 48 |  | PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-2 | -6.132e+00 | 0.0111 | 929.0 | 37.20% | 337.9 | 34.55% | motif file (matrix) |
pdf |
| 49 |  | NPAS2(HLH)/Liver-NPAS2-ChIP-Seq(GSE39860)/Homer | 1e-2 | -6.094e+00 | 0.0113 | 667.0 | 26.71% | 237.7 | 24.30% | motif file (matrix) |
pdf |
| 50 |  | FXR(NR/IR1)/Liver-FXR-ChIP-Seq(Chong et al.)/Homer | 1e-2 | -5.958e+00 | 0.0127 | 191.0 | 7.65% | 61.0 | 6.24% | motif file (matrix) |
pdf |
| 51 |  | HNF4a(NR/DR1)/HepG2-HNF4a-ChIP-Seq(GSE25021)/Homer | 1e-2 | -5.901e+00 | 0.0132 | 252.0 | 10.09% | 83.6 | 8.55% | motif file (matrix) |
pdf |
| 52 |  | ERE(NR/IR3)/MCF7-ERa-ChIP-Seq(Unpublished)/Homer | 1e-2 | -5.699e+00 | 0.0158 | 139.0 | 5.57% | 43.6 | 4.46% | motif file (matrix) |
pdf |
| 53 | 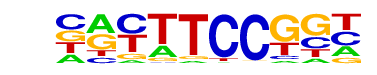 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-2 | -5.535e+00 | 0.0183 | 552.0 | 22.11% | 195.2 | 19.96% | motif file (matrix) |
pdf |
| 54 |  | RUNX2(Runt)/PCa-RUNX2-ChIP-Seq(GSE33889)/Homer | 1e-2 | -5.441e+00 | 0.0197 | 554.0 | 22.19% | 196.7 | 20.11% | motif file (matrix) |
pdf |
| 55 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-2 | -5.419e+00 | 0.0198 | 874.0 | 35.00% | 318.1 | 32.52% | motif file (matrix) |
pdf |
| 56 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-2 | -5.347e+00 | 0.0209 | 323.0 | 12.94% | 110.5 | 11.30% | motif file (matrix) |
pdf |
| 57 |  | Egr1(Zf)/K562-Egr1-ChIP-Seq(GSE32465)/Homer | 1e-2 | -5.296e+00 | 0.0216 | 227.0 | 9.09% | 75.8 | 7.75% | motif file (matrix) |
pdf |
| 58 |  | LXRE(NR/DR4)/BLRP(RAW)-LXRb-ChIP-Seq(GSE21512)/Homer | 1e-2 | -5.161e+00 | 0.0243 | 168.0 | 6.73% | 54.7 | 5.60% | motif file (matrix) |
pdf |
| 59 |  | TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer | 1e-2 | -4.991e+00 | 0.0284 | 564.0 | 22.59% | 202.0 | 20.65% | motif file (matrix) |
pdf |
| 60 |  | NFAT(RHD)/Jurkat-NFATC1-ChIP-Seq(Jolma et al.)/Homer | 1e-2 | -4.906e+00 | 0.0303 | 776.0 | 31.08% | 282.0 | 28.83% | motif file (matrix) |
pdf |
| 61 |  | Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer | 1e-2 | -4.894e+00 | 0.0303 | 178.0 | 7.13% | 58.8 | 6.01% | motif file (matrix) |
pdf |
| 62 |  | HIF-1a(HLH)/MCF7-HIF1a-ChIP-Seq(GSE28352)/Homer | 1e-2 | -4.825e+00 | 0.0318 | 130.0 | 5.21% | 41.9 | 4.28% | motif file (matrix) |
pdf |
| 63 |  | E-box(HLH)/Promoter/Homer | 1e-2 | -4.745e+00 | 0.0340 | 19.0 | 0.76% | 4.2 | 0.43% | motif file (matrix) |
pdf |
| 64 |  | GATA-IR4(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-2 | -4.744e+00 | 0.0340 | 75.0 | 3.00% | 22.1 | 2.26% | motif file (matrix) |
pdf |
| 65 |  | Unknown-ESC-element/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-2 | -4.721e+00 | 0.0340 | 276.0 | 11.05% | 94.7 | 9.68% | motif file (matrix) |
pdf |
| 66 |  | BORIS(Zf)/K562-CTCFL-ChIP-Seq(GSE32465)/Homer | 1e-2 | -4.699e+00 | 0.0340 | 54.0 | 2.16% | 15.6 | 1.59% | motif file (matrix) |
pdf |
| 67 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-2 | -4.629e+00 | 0.0359 | 450.0 | 18.02% | 159.4 | 16.30% | motif file (matrix) |
pdf |





































































































































