| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | CTCF(Zf)/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-57 | -1.331e+02 | 0.0000 | 92.0 | 10.91% | 8.6 | 1.22% | motif file (matrix) |
pdf |
| 2 |  | BORIS(Zf)/K562-CTCFL-ChIP-Seq(GSE32465)/Homer | 1e-53 | -1.233e+02 | 0.0000 | 96.0 | 11.39% | 10.2 | 1.44% | motif file (matrix) |
pdf |
| 3 |  | Hnf1(Homeobox)/Liver-Foxa2-Chip-Seq(GSE25694)/Homer | 1e-40 | -9.212e+01 | 0.0000 | 51.0 | 6.05% | 3.2 | 0.46% | motif file (matrix) |
pdf |
| 4 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-22 | -5.275e+01 | 0.0000 | 76.0 | 9.02% | 16.9 | 2.39% | motif file (matrix) |
pdf |
| 5 |  | GATA3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-22 | -5.178e+01 | 0.0000 | 206.0 | 24.44% | 85.1 | 12.06% | motif file (matrix) |
pdf |
| 6 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-19 | -4.394e+01 | 0.0000 | 79.0 | 9.37% | 20.3 | 2.88% | motif file (matrix) |
pdf |
| 7 |  | Rfx2(HTH)/LoVo-RFX2-ChIP-Seq(GSE49402)/Homer | 1e-18 | -4.299e+01 | 0.0000 | 21.0 | 2.49% | 1.9 | 0.27% | motif file (matrix) |
pdf |
| 8 |  | Erra(NR)/HepG2-Erra-ChIP-Seq(GSE31477)/Homer | 1e-17 | -4.137e+01 | 0.0000 | 283.0 | 33.57% | 145.2 | 20.58% | motif file (matrix) |
pdf |
| 9 |  | Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-17 | -4.091e+01 | 0.0000 | 41.0 | 4.86% | 6.1 | 0.86% | motif file (matrix) |
pdf |
| 10 |  | Rfx1(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-16 | -3.768e+01 | 0.0000 | 36.0 | 4.27% | 5.8 | 0.82% | motif file (matrix) |
pdf |
| 11 | 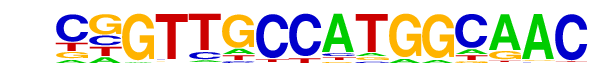 | RFX(HTH)/K562-RFX3-ChIP-Seq(SRA012198)/Homer | 1e-16 | -3.726e+01 | 0.0000 | 19.0 | 2.25% | 1.1 | 0.16% | motif file (matrix) |
pdf |
| 12 |  | Gata1(Zf)/K562-GATA1-ChIP-Seq(GSE18829)/Homer | 1e-15 | -3.539e+01 | 0.0000 | 109.0 | 12.93% | 39.3 | 5.57% | motif file (matrix) |
pdf |
| 13 |  | Fox:Ebox(Forkhead:HLH)/Panc1-Foxa2-ChIP-Seq(GSE47459)/Homer | 1e-15 | -3.525e+01 | 0.0000 | 130.0 | 15.42% | 51.5 | 7.30% | motif file (matrix) |
pdf |
| 14 |  | Gata2(Zf)/K562-GATA2-ChIP-Seq(GSE18829)/Homer | 1e-14 | -3.431e+01 | 0.0000 | 115.0 | 13.64% | 43.8 | 6.21% | motif file (matrix) |
pdf |
| 15 |  | FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-14 | -3.405e+01 | 0.0000 | 132.0 | 15.66% | 53.7 | 7.62% | motif file (matrix) |
pdf |
| 16 |  | AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-14 | -3.393e+01 | 0.0000 | 81.0 | 9.61% | 25.5 | 3.62% | motif file (matrix) |
pdf |
| 17 |  | Foxa2(Forkhead)/Liver-Foxa2-ChIP-Seq(GSE25694)/Homer | 1e-14 | -3.301e+01 | 0.0000 | 110.0 | 13.05% | 41.5 | 5.89% | motif file (matrix) |
pdf |
| 18 |  | Gata4(Zf)/Heart-Gata4-ChIP-Seq(GSE35151)/Homer | 1e-14 | -3.258e+01 | 0.0000 | 150.0 | 17.79% | 65.4 | 9.26% | motif file (matrix) |
pdf |
| 19 |  | FOXA1(Forkhead)/MCF7-FOXA1-ChIP-Seq(GSE26831)/Homer | 1e-13 | -3.123e+01 | 0.0000 | 108.0 | 12.81% | 41.0 | 5.81% | motif file (matrix) |
pdf |
| 20 |  | HNF4a(NR/DR1)/HepG2-HNF4a-ChIP-Seq(GSE25021)/Homer | 1e-13 | -3.057e+01 | 0.0000 | 109.0 | 12.93% | 42.3 | 6.00% | motif file (matrix) |
pdf |
| 21 |  | Tcf3(HMG)/mES-Tcf3-ChIP-Seq(GSE11724)/Homer | 1e-12 | -2.932e+01 | 0.0000 | 37.0 | 4.39% | 7.7 | 1.09% | motif file (matrix) |
pdf |
| 22 |  | Tcf4(HMG)/Hct116-Tcf4-ChIP-Seq(SRA012054)/Homer | 1e-12 | -2.894e+01 | 0.0000 | 58.0 | 6.88% | 16.9 | 2.40% | motif file (matrix) |
pdf |
| 23 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-12 | -2.778e+01 | 0.0000 | 57.0 | 6.76% | 16.9 | 2.39% | motif file (matrix) |
pdf |
| 24 |  | Nkx6.1(Homeobox)/Islet-Nkx6.1-ChIP-Seq(GSE40975)/Homer | 1e-11 | -2.664e+01 | 0.0000 | 221.0 | 26.22% | 118.3 | 16.77% | motif file (matrix) |
pdf |
| 25 |  | Hoxb4(Homeobox)/ES-Hoxb4-ChIP-Seq(GSE34014)/Homer | 1e-11 | -2.542e+01 | 0.0000 | 26.0 | 3.08% | 4.7 | 0.67% | motif file (matrix) |
pdf |
| 26 |  | RXR(NR/DR1)/3T3L1-RXR-ChIP-Seq(GSE13511)/Homer | 1e-9 | -2.225e+01 | 0.0000 | 140.0 | 16.61% | 68.1 | 9.65% | motif file (matrix) |
pdf |
| 27 |  | PPARE(NR/DR1)/3T3L1-Pparg-ChIP-Seq(GSE13511)/Homer | 1e-8 | -1.961e+01 | 0.0000 | 122.0 | 14.47% | 59.6 | 8.45% | motif file (matrix) |
pdf |
| 28 |  | TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer | 1e-8 | -1.945e+01 | 0.0000 | 85.0 | 10.08% | 36.5 | 5.17% | motif file (matrix) |
pdf |
| 29 |  | Lhx2(Homeobox)/HFSC-Lhx2-ChIP-Seq(GSE48068)/Homer | 1e-8 | -1.882e+01 | 0.0000 | 102.0 | 12.10% | 47.4 | 6.72% | motif file (matrix) |
pdf |
| 30 |  | TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer | 1e-7 | -1.774e+01 | 0.0000 | 122.0 | 14.47% | 62.0 | 8.79% | motif file (matrix) |
pdf |
| 31 |  | Sox6(HMG)/Myotubes-Sox6-ChIP-Seq(GSE32627)/Homer | 1e-7 | -1.739e+01 | 0.0000 | 174.0 | 20.64% | 97.3 | 13.79% | motif file (matrix) |
pdf |
| 32 |  | Foxo1(Forkhead)/RAW-Foxo1-ChIP-Seq(Fan et al.)/Homer | 1e-7 | -1.692e+01 | 0.0000 | 201.0 | 23.84% | 117.7 | 16.68% | motif file (matrix) |
pdf |
| 33 | 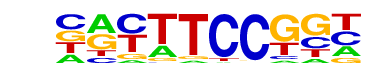 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-7 | -1.657e+01 | 0.0000 | 123.0 | 14.59% | 63.1 | 8.94% | motif file (matrix) |
pdf |
| 34 |  | X-box(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-6 | -1.569e+01 | 0.0000 | 20.0 | 2.37% | 4.3 | 0.61% | motif file (matrix) |
pdf |
| 35 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-6 | -1.518e+01 | 0.0000 | 104.0 | 12.34% | 52.6 | 7.45% | motif file (matrix) |
pdf |
| 36 |  | Eomes(T-box)/H9-Eomes-ChIP-Seq(GSE26097)/Homer | 1e-6 | -1.500e+01 | 0.0000 | 209.0 | 24.79% | 126.2 | 17.88% | motif file (matrix) |
pdf |
| 37 |  | USF1(HLH)/GM12878-Usf1-ChIP-Seq(GSE32465)/Homer | 1e-6 | -1.468e+01 | 0.0000 | 55.0 | 6.52% | 22.9 | 3.24% | motif file (matrix) |
pdf |
| 38 |  | NF-E2(bZIP)/K562-NFE2-ChIP-Seq(GSE31477)/Homer | 1e-6 | -1.444e+01 | 0.0000 | 10.0 | 1.19% | 1.6 | 0.22% | motif file (matrix) |
pdf |
| 39 |  | NFkB-p50,p52(RHD)/p50-ChIP-Chip(Schreiber et al.)/Homer | 1e-6 | -1.444e+01 | 0.0000 | 10.0 | 1.19% | 1.7 | 0.24% | motif file (matrix) |
pdf |
| 40 |  | bHLHE40(HLH)/HepG2-BHLHE40-ChIP-Seq(GSE31477)/Homer | 1e-6 | -1.392e+01 | 0.0000 | 34.0 | 4.03% | 11.6 | 1.65% | motif file (matrix) |
pdf |
| 41 |  | c-Jun-CRE(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-5 | -1.354e+01 | 0.0000 | 43.0 | 5.10% | 17.0 | 2.41% | motif file (matrix) |
pdf |
| 42 |  | Sox3(HMG)/NPC-Sox3-ChIP-Seq(GSE33059)/Homer | 1e-5 | -1.329e+01 | 0.0000 | 190.0 | 22.54% | 115.2 | 16.32% | motif file (matrix) |
pdf |
| 43 |  | TR4(NR/DR1)/Hela-TR4-ChIP-Seq(GSE24685)/Homer | 1e-5 | -1.292e+01 | 0.0000 | 27.0 | 3.20% | 8.5 | 1.20% | motif file (matrix) |
pdf |
| 44 |  | Sox2(HMG)/mES-Sox2-ChIP-Seq(GSE11431)/Homer | 1e-5 | -1.285e+01 | 0.0000 | 119.0 | 14.12% | 65.9 | 9.34% | motif file (matrix) |
pdf |
| 45 |  | Oct4:Sox17/F9-Sox17-ChIP-Seq(GSE44553)/Homer | 1e-5 | -1.215e+01 | 0.0000 | 15.0 | 1.78% | 3.9 | 0.55% | motif file (matrix) |
pdf |
| 46 |  | Nur77(NR)/K562-NR4A1-ChIP-Seq(GSE31363)/Homer | 1e-5 | -1.197e+01 | 0.0000 | 32.0 | 3.80% | 11.5 | 1.63% | motif file (matrix) |
pdf |
| 47 |  | Lhx3(Homeobox)/Neuron-Lhx3-ChIP-Seq(GSE31456)/Homer | 1e-5 | -1.181e+01 | 0.0000 | 141.0 | 16.73% | 83.0 | 11.76% | motif file (matrix) |
pdf |
| 48 |  | MafA(bZIP)/Islet-MafA-ChIP-Seq(GSE30298)/Homer | 1e-5 | -1.157e+01 | 0.0000 | 93.0 | 11.03% | 49.2 | 6.98% | motif file (matrix) |
pdf |
| 49 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-4 | -1.107e+01 | 0.0001 | 111.0 | 13.17% | 63.0 | 8.93% | motif file (matrix) |
pdf |
| 50 |  | Mef2a(MADS)/HL1-Mef2a.biotin-ChIP-Seq(GSE21529/Homer | 1e-4 | -1.081e+01 | 0.0001 | 25.0 | 2.97% | 8.9 | 1.26% | motif file (matrix) |
pdf |
| 51 |  | E2F(E2F)/Cell-Cycle-Exp/Homer | 1e-4 | -1.025e+01 | 0.0002 | 8.0 | 0.95% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 52 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-4 | -9.827e+00 | 0.0003 | 173.0 | 20.52% | 109.1 | 15.47% | motif file (matrix) |
pdf |
| 53 |  | CArG(MADS)/PUER-Srf-ChIP-Seq(Sullivan et al.)/Homer | 1e-4 | -9.787e+00 | 0.0003 | 20.0 | 2.37% | 6.1 | 0.87% | motif file (matrix) |
pdf |
| 54 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-4 | -9.707e+00 | 0.0003 | 67.0 | 7.95% | 34.9 | 4.95% | motif file (matrix) |
pdf |
| 55 |  | NFY(CCAAT)/Promoter/Homer | 1e-4 | -9.678e+00 | 0.0003 | 73.0 | 8.66% | 38.9 | 5.51% | motif file (matrix) |
pdf |
| 56 |  | Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer | 1e-4 | -9.510e+00 | 0.0003 | 38.0 | 4.51% | 16.4 | 2.32% | motif file (matrix) |
pdf |
| 57 |  | Usf2(HLH)/C2C12-Usf2-ChIP-Seq(GSE36030)/Homer | 1e-4 | -9.510e+00 | 0.0003 | 38.0 | 4.51% | 16.4 | 2.32% | motif file (matrix) |
pdf |
| 58 |  | TRa(NR)/C17.2-TRa-ChIP-Seq(GSE38347)/Homer | 1e-4 | -9.340e+00 | 0.0004 | 54.0 | 6.41% | 26.4 | 3.74% | motif file (matrix) |
pdf |
| 59 |  | TCFL2(HMG)/K562-TCF7L2-ChIP-Seq(GSE29196)/Homer | 1e-3 | -8.761e+00 | 0.0007 | 17.0 | 2.02% | 5.1 | 0.72% | motif file (matrix) |
pdf |
| 60 |  | Bach2(bZIP)/OCILy7-Bach2-ChIP-Seq(GSE44420)/Homer | 1e-3 | -8.760e+00 | 0.0007 | 21.0 | 2.49% | 7.4 | 1.05% | motif file (matrix) |
pdf |
| 61 | 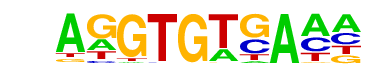 | Tbet(T-box)/CD8-Tbet-ChIP-Seq(GSE33802)/Homer | 1e-3 | -8.337e+00 | 0.0010 | 109.0 | 12.93% | 65.9 | 9.34% | motif file (matrix) |
pdf |
| 62 |  | Nrf2(bZIP)/Lymphoblast-Nrf2-ChIP-Seq(GSE37589)/Homer | 1e-3 | -8.328e+00 | 0.0010 | 7.0 | 0.83% | 1.2 | 0.17% | motif file (matrix) |
pdf |
| 63 |  | n-Myc(HLH)/mES-nMyc-ChIP-Seq(GSE11431)/Homer | 1e-3 | -8.098e+00 | 0.0012 | 58.0 | 6.88% | 31.0 | 4.39% | motif file (matrix) |
pdf |
| 64 |  | MafK(bZIP)/C2C12-MafK-ChIP-Seq(GSE36030)/Homer | 1e-3 | -8.082e+00 | 0.0012 | 24.0 | 2.85% | 9.7 | 1.38% | motif file (matrix) |
pdf |
| 65 |  | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-3 | -8.021e+00 | 0.0012 | 122.0 | 14.47% | 75.8 | 10.75% | motif file (matrix) |
pdf |
| 66 |  | PU.1(ETS)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-3 | -7.951e+00 | 0.0013 | 47.0 | 5.58% | 23.2 | 3.29% | motif file (matrix) |
pdf |
| 67 |  | EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer | 1e-3 | -7.619e+00 | 0.0018 | 66.0 | 7.83% | 36.2 | 5.13% | motif file (matrix) |
pdf |
| 68 |  | Esrrb(NR)/mES-Esrrb-ChIP-Seq(GSE11431)/Homer | 1e-3 | -7.437e+00 | 0.0021 | 70.0 | 8.30% | 39.2 | 5.56% | motif file (matrix) |
pdf |
| 69 |  | c-Myc(HLH)/LNCAP-cMyc-ChIP-Seq(unpublished)/Homer | 1e-3 | -7.002e+00 | 0.0032 | 36.0 | 4.27% | 17.1 | 2.42% | motif file (matrix) |
pdf |
| 70 |  | CLOCK(HLH)/Liver-Clock-ChIP-Seq(GSE39860)/Homer | 1e-2 | -6.783e+00 | 0.0040 | 51.0 | 6.05% | 27.8 | 3.95% | motif file (matrix) |
pdf |
| 71 |  | Znf263(Zf)/K562-Znf263-ChIP-Seq(GSE31477)/Homer | 1e-2 | -6.657e+00 | 0.0045 | 233.0 | 27.64% | 163.6 | 23.18% | motif file (matrix) |
pdf |
| 72 |  | CTCF-SatelliteElement/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-2 | -6.531e+00 | 0.0049 | 6.0 | 0.71% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 73 |  | PAX5-shortForm(Paired/Homeobox)/GM12878-PAX5-ChIP-Seq(GSE32465)/Homer | 1e-2 | -6.531e+00 | 0.0049 | 6.0 | 0.71% | 1.6 | 0.23% | motif file (matrix) |
pdf |
| 74 |  | Unknown-ESC-element/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-2 | -6.433e+00 | 0.0053 | 93.0 | 11.03% | 57.3 | 8.12% | motif file (matrix) |
pdf |
| 75 |  | Atf1(bZIP)/K562-ATF1-ChIP-Seq(GSE31477)/Homer | 1e-2 | -6.350e+00 | 0.0057 | 69.0 | 8.19% | 40.2 | 5.70% | motif file (matrix) |
pdf |
| 76 |  | AP-2alpha(AP2)/Hela-AP2alpha-ChIP-Seq(GSE31477)/Homer | 1e-2 | -6.349e+00 | 0.0057 | 101.0 | 11.98% | 63.4 | 8.99% | motif file (matrix) |
pdf |
| 77 | 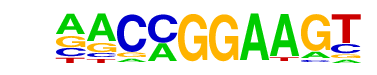 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-2 | -6.073e+00 | 0.0074 | 143.0 | 16.96% | 95.8 | 13.58% | motif file (matrix) |
pdf |
| 78 |  | AP-2gamma(AP2)/MCF7-TFAP2C-ChIP-Seq(GSE21234)/Homer | 1e-2 | -6.037e+00 | 0.0075 | 123.0 | 14.59% | 80.1 | 11.35% | motif file (matrix) |
pdf |
| 79 |  | HIF-1a(HLH)/MCF7-HIF1a-ChIP-Seq(GSE28352)/Homer | 1e-2 | -6.020e+00 | 0.0076 | 18.0 | 2.14% | 7.2 | 1.03% | motif file (matrix) |
pdf |
| 80 |  | Klf4(Zf)/mES-Klf4-ChIP-Seq(GSE11431)/Homer | 1e-2 | -5.759e+00 | 0.0097 | 46.0 | 5.46% | 25.7 | 3.65% | motif file (matrix) |
pdf |
| 81 |  | Bach1(bZIP)/K562-Bach1-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.740e+00 | 0.0097 | 8.0 | 0.95% | 2.4 | 0.35% | motif file (matrix) |
pdf |
| 82 |  | HOXA2(Homeobox)/mES-Hoxa2-ChIP-Seq(Donaldson et al.)/Homer | 1e-2 | -5.740e+00 | 0.0097 | 8.0 | 0.95% | 2.7 | 0.39% | motif file (matrix) |
pdf |
| 83 |  | Chop(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer | 1e-2 | -5.580e+00 | 0.0112 | 12.0 | 1.42% | 4.6 | 0.65% | motif file (matrix) |
pdf |
| 84 |  | GATA-IR3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-2 | -5.562e+00 | 0.0113 | 10.0 | 1.19% | 3.5 | 0.49% | motif file (matrix) |
pdf |
| 85 |  | GATA:SCL/Ter119-SCL-ChIP-Seq(GSE18720)/Homer | 1e-2 | -5.204e+00 | 0.0159 | 22.0 | 2.61% | 10.9 | 1.55% | motif file (matrix) |
pdf |
| 86 |  | Foxh1(Forkhead)/hESC-FOXH1-ChIP-Seq(GSE29422)/Homer | 1e-2 | -4.986e+00 | 0.0195 | 40.0 | 4.74% | 22.8 | 3.23% | motif file (matrix) |
pdf |
| 87 |  | MYB(HTH)/ERMYB-Myb-ChIPSeq(GSE22095)/Homer | 1e-2 | -4.957e+00 | 0.0199 | 152.0 | 18.03% | 105.2 | 14.91% | motif file (matrix) |
pdf |
| 88 |  | STAT1(Stat)/HelaS3-STAT1-ChIP-Seq(GSE12782)/Homer | 1e-2 | -4.949e+00 | 0.0199 | 20.0 | 2.37% | 9.8 | 1.38% | motif file (matrix) |
pdf |
| 89 |  | REST-NRSF(Zf)/Jurkat-NRSF-ChIP-Seq/Homer | 1e-2 | -4.882e+00 | 0.0210 | 5.0 | 0.59% | 0.8 | 0.11% | motif file (matrix) |
pdf |
| 90 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-2 | -4.870e+00 | 0.0210 | 47.0 | 5.58% | 27.7 | 3.93% | motif file (matrix) |
pdf |
| 91 |  | BMAL1(HLH)/Liver-Bmal1-ChIP-Seq(GSE39860)/Homer | 1e-2 | -4.662e+00 | 0.0255 | 134.0 | 15.90% | 92.4 | 13.10% | motif file (matrix) |
pdf |





















































































































































































