| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-879 | -2.026e+03 | 0.0000 | 5197.0 | 20.13% | 1969.0 | 7.68% | motif file (matrix) |
pdf |
| 2 |  | FOXA1(Forkhead)/MCF7-FOXA1-ChIP-Seq(GSE26831)/Homer | 1e-841 | -1.937e+03 | 0.0000 | 4414.0 | 17.10% | 1538.3 | 6.00% | motif file (matrix) |
pdf |
| 3 |  | Foxa2(Forkhead)/Liver-Foxa2-ChIP-Seq(GSE25694)/Homer | 1e-646 | -1.489e+03 | 0.0000 | 3809.0 | 14.76% | 1413.3 | 5.52% | motif file (matrix) |
pdf |
| 4 |  | Fox:Ebox(Forkhead:HLH)/Panc1-Foxa2-ChIP-Seq(GSE47459)/Homer | 1e-559 | -1.289e+03 | 0.0000 | 4497.0 | 17.42% | 1984.7 | 7.75% | motif file (matrix) |
pdf |
| 5 |  | Gata4(Zf)/Heart-Gata4-ChIP-Seq(GSE35151)/Homer | 1e-542 | -1.249e+03 | 0.0000 | 4307.0 | 16.69% | 1885.2 | 7.36% | motif file (matrix) |
pdf |
| 6 |  | CTCF(Zf)/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-541 | -1.247e+03 | 0.0000 | 1556.0 | 6.03% | 321.8 | 1.26% | motif file (matrix) |
pdf |
| 7 |  | Gata2(Zf)/K562-GATA2-ChIP-Seq(GSE18829)/Homer | 1e-501 | -1.155e+03 | 0.0000 | 3172.0 | 12.29% | 1216.8 | 4.75% | motif file (matrix) |
pdf |
| 8 |  | GATA3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-498 | -1.148e+03 | 0.0000 | 5545.0 | 21.48% | 2845.8 | 11.11% | motif file (matrix) |
pdf |
| 9 |  | Gata1(Zf)/K562-GATA1-ChIP-Seq(GSE18829)/Homer | 1e-489 | -1.127e+03 | 0.0000 | 2924.0 | 11.33% | 1081.4 | 4.22% | motif file (matrix) |
pdf |
| 10 |  | BORIS(Zf)/K562-CTCFL-ChIP-Seq(GSE32465)/Homer | 1e-444 | -1.023e+03 | 0.0000 | 1866.0 | 7.23% | 538.7 | 2.10% | motif file (matrix) |
pdf |
| 11 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-307 | -7.091e+02 | 0.0000 | 1878.0 | 7.28% | 694.2 | 2.71% | motif file (matrix) |
pdf |
| 12 |  | Unknown-ESC-element/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-255 | -5.892e+02 | 0.0000 | 3162.0 | 12.25% | 1646.9 | 6.43% | motif file (matrix) |
pdf |
| 13 |  | Lhx3(Homeobox)/Neuron-Lhx3-ChIP-Seq(GSE31456)/Homer | 1e-245 | -5.647e+02 | 0.0000 | 4900.0 | 18.98% | 3014.0 | 11.76% | motif file (matrix) |
pdf |
| 14 |  | Lhx2(Homeobox)/HFSC-Lhx2-ChIP-Seq(GSE48068)/Homer | 1e-230 | -5.305e+02 | 0.0000 | 3341.0 | 12.94% | 1842.7 | 7.19% | motif file (matrix) |
pdf |
| 15 |  | Sox3(HMG)/NPC-Sox3-ChIP-Seq(GSE33059)/Homer | 1e-229 | -5.282e+02 | 0.0000 | 5772.0 | 22.36% | 3785.9 | 14.78% | motif file (matrix) |
pdf |
| 16 |  | Sox6(HMG)/Myotubes-Sox6-ChIP-Seq(GSE32627)/Homer | 1e-221 | -5.104e+02 | 0.0000 | 4949.0 | 19.17% | 3133.8 | 12.23% | motif file (matrix) |
pdf |
| 17 |  | Sox2(HMG)/mES-Sox2-ChIP-Seq(GSE11431)/Homer | 1e-215 | -4.963e+02 | 0.0000 | 3223.0 | 12.49% | 1793.1 | 7.00% | motif file (matrix) |
pdf |
| 18 |  | Eomes(T-box)/H9-Eomes-ChIP-Seq(GSE26097)/Homer | 1e-199 | -4.591e+02 | 0.0000 | 6699.0 | 25.95% | 4700.6 | 18.35% | motif file (matrix) |
pdf |
| 19 |  | Nkx6.1(Homeobox)/Islet-Nkx6.1-ChIP-Seq(GSE40975)/Homer | 1e-192 | -4.441e+02 | 0.0000 | 6719.0 | 26.03% | 4747.4 | 18.53% | motif file (matrix) |
pdf |
| 20 |  | Foxo1(Forkhead)/RAW-Foxo1-ChIP-Seq(Fan et al.)/Homer | 1e-182 | -4.191e+02 | 0.0000 | 6089.0 | 23.59% | 4250.1 | 16.59% | motif file (matrix) |
pdf |
| 21 |  | CRX(Homeobox)/Retina-Crx-ChIP-Seq(GSE20012)/Homer | 1e-155 | -3.590e+02 | 0.0000 | 6958.0 | 26.96% | 5140.7 | 20.06% | motif file (matrix) |
pdf |
| 22 |  | Nanog(Homeobox)/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-133 | -3.076e+02 | 0.0000 | 11033.0 | 42.75% | 9048.4 | 35.32% | motif file (matrix) |
pdf |
| 23 | 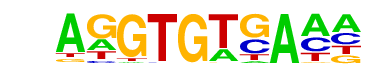 | Tbet(T-box)/CD8-Tbet-ChIP-Seq(GSE33802)/Homer | 1e-128 | -2.955e+02 | 0.0000 | 3276.0 | 12.69% | 2116.7 | 8.26% | motif file (matrix) |
pdf |
| 24 |  | Tbx5(T-box)/HL1-Tbx5.biotin-ChIP-Seq(GSE21529)/Homer | 1e-127 | -2.938e+02 | 0.0000 | 9999.0 | 38.74% | 8108.8 | 31.65% | motif file (matrix) |
pdf |
| 25 |  | Isl1(Homeobox)/Neuron-Isl1-ChIP-Seq(GSE31456)/Homer | 1e-108 | -2.509e+02 | 0.0000 | 5505.0 | 21.33% | 4111.9 | 16.05% | motif file (matrix) |
pdf |
| 26 |  | Tcf4(HMG)/Hct116-Tcf4-ChIP-Seq(SRA012054)/Homer | 1e-94 | -2.173e+02 | 0.0000 | 1484.0 | 5.75% | 828.1 | 3.23% | motif file (matrix) |
pdf |
| 27 |  | Pitx1(Homeobox)/Chicken-Pitx1-ChIP-Seq(GSE38910)/Homer | 1e-68 | -1.574e+02 | 0.0000 | 10700.0 | 41.46% | 9265.9 | 36.16% | motif file (matrix) |
pdf |
| 28 |  | TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer | 1e-66 | -1.532e+02 | 0.0000 | 2381.0 | 9.22% | 1646.3 | 6.43% | motif file (matrix) |
pdf |
| 29 |  | Tcf3(HMG)/mES-Tcf3-ChIP-Seq(GSE11724)/Homer | 1e-66 | -1.526e+02 | 0.0000 | 906.0 | 3.51% | 481.6 | 1.88% | motif file (matrix) |
pdf |
| 30 |  | KLF5(Zf)/LoVo-KLF5-ChIP-Seq(GSE49402)/Homer | 1e-64 | -1.492e+02 | 0.0000 | 5021.0 | 19.45% | 3968.3 | 15.49% | motif file (matrix) |
pdf |
| 31 | 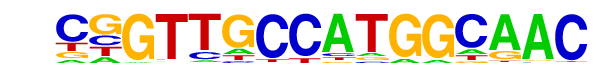 | RFX(HTH)/K562-RFX3-ChIP-Seq(SRA012198)/Homer | 1e-62 | -1.442e+02 | 0.0000 | 486.0 | 1.88% | 203.4 | 0.79% | motif file (matrix) |
pdf |
| 32 |  | Sp1(Zf)/Promoter/Homer | 1e-57 | -1.327e+02 | 0.0000 | 1503.0 | 5.82% | 966.6 | 3.77% | motif file (matrix) |
pdf |
| 33 |  | X-box(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-57 | -1.321e+02 | 0.0000 | 500.0 | 1.94% | 221.6 | 0.86% | motif file (matrix) |
pdf |
| 34 |  | Rfx2(HTH)/LoVo-RFX2-ChIP-Seq(GSE49402)/Homer | 1e-54 | -1.251e+02 | 0.0000 | 502.0 | 1.94% | 228.5 | 0.89% | motif file (matrix) |
pdf |
| 35 |  | SCL(HLH)/HPC7-Scl-ChIP-Seq(GSE13511)/Homer | 1e-51 | -1.188e+02 | 0.0000 | 13464.0 | 52.16% | 12155.3 | 47.44% | motif file (matrix) |
pdf |
| 36 |  | TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer | 1e-49 | -1.133e+02 | 0.0000 | 1704.0 | 6.60% | 1167.8 | 4.56% | motif file (matrix) |
pdf |
| 37 |  | Rfx1(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-47 | -1.100e+02 | 0.0000 | 794.0 | 3.08% | 450.1 | 1.76% | motif file (matrix) |
pdf |
| 38 |  | NFY(CCAAT)/Promoter/Homer | 1e-44 | -1.013e+02 | 0.0000 | 2289.0 | 8.87% | 1691.9 | 6.60% | motif file (matrix) |
pdf |
| 39 |  | Maz(Zf)/HepG2-Maz-ChIP-Seq(GSE31477)/Homer | 1e-42 | -9.747e+01 | 0.0000 | 5306.0 | 20.56% | 4421.5 | 17.26% | motif file (matrix) |
pdf |
| 40 |  | Oct4(POU/Homeobox)/mES-Oct4-ChIP-Seq(GSE11431)/Homer | 1e-41 | -9.621e+01 | 0.0000 | 998.0 | 3.87% | 627.1 | 2.45% | motif file (matrix) |
pdf |
| 41 |  | Hnf1(Homeobox)/Liver-Foxa2-Chip-Seq(GSE25694)/Homer | 1e-41 | -9.585e+01 | 0.0000 | 377.0 | 1.46% | 170.7 | 0.67% | motif file (matrix) |
pdf |
| 42 |  | Foxh1(Forkhead)/hESC-FOXH1-ChIP-Seq(GSE29422)/Homer | 1e-38 | -8.884e+01 | 0.0000 | 1395.0 | 5.40% | 963.5 | 3.76% | motif file (matrix) |
pdf |
| 43 |  | Klf4(Zf)/mES-Klf4-ChIP-Seq(GSE11431)/Homer | 1e-38 | -8.833e+01 | 0.0000 | 1504.0 | 5.83% | 1055.3 | 4.12% | motif file (matrix) |
pdf |
| 44 |  | Ptf1a(HLH)/Panc1-Ptf1a-ChIP-Seq(GSE47459)/Homer | 1e-35 | -8.108e+01 | 0.0000 | 8105.0 | 31.40% | 7143.3 | 27.88% | motif file (matrix) |
pdf |
| 45 |  | E2A-nearPU.1(HLH)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-35 | -8.095e+01 | 0.0000 | 4517.0 | 17.50% | 3763.7 | 14.69% | motif file (matrix) |
pdf |
| 46 |  | Oct2(POU/Homeobox)/Bcell-Oct2-ChIP-Seq(GSE21512)/Homer | 1e-34 | -8.005e+01 | 0.0000 | 594.0 | 2.30% | 340.7 | 1.33% | motif file (matrix) |
pdf |
| 47 |  | HNF6(Homeobox)/Liver-Hnf6-ChIP-Seq(ERP000394)/Homer | 1e-34 | -7.906e+01 | 0.0000 | 1102.0 | 4.27% | 742.8 | 2.90% | motif file (matrix) |
pdf |
| 48 |  | Phox2a(Homeobox)/Neuron-Phox2a-ChIP-Seq(GSE31456)/Homer | 1e-34 | -7.886e+01 | 0.0000 | 772.0 | 2.99% | 478.4 | 1.87% | motif file (matrix) |
pdf |
| 49 |  | REST-NRSF(Zf)/Jurkat-NRSF-ChIP-Seq/Homer | 1e-33 | -7.648e+01 | 0.0000 | 98.0 | 0.38% | 21.5 | 0.08% | motif file (matrix) |
pdf |
| 50 |  | TCFL2(HMG)/K562-TCF7L2-ChIP-Seq(GSE29196)/Homer | 1e-32 | -7.591e+01 | 0.0000 | 348.0 | 1.35% | 168.0 | 0.66% | motif file (matrix) |
pdf |
| 51 |  | E2A(HLH)/proBcell-E2A-ChIP-Seq(GSE21978)/Homer | 1e-32 | -7.462e+01 | 0.0000 | 4887.0 | 18.93% | 4135.9 | 16.14% | motif file (matrix) |
pdf |
| 52 |  | CTCF-SatelliteElement/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-30 | -7.101e+01 | 0.0000 | 99.0 | 0.38% | 23.1 | 0.09% | motif file (matrix) |
pdf |
| 53 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-30 | -7.047e+01 | 0.0000 | 1495.0 | 5.79% | 1091.5 | 4.26% | motif file (matrix) |
pdf |
| 54 |  | Tcf12(HLH)/GM12878-Tcf12-ChIP-Seq(GSE32465)/Homer | 1e-28 | -6.503e+01 | 0.0000 | 3446.0 | 13.35% | 2848.4 | 11.12% | motif file (matrix) |
pdf |
| 55 |  | NF1:FOXA1/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-28 | -6.490e+01 | 0.0000 | 204.0 | 0.79% | 83.1 | 0.32% | motif file (matrix) |
pdf |
| 56 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-25 | -5.911e+01 | 0.0000 | 3127.0 | 12.11% | 2582.6 | 10.08% | motif file (matrix) |
pdf |
| 57 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-25 | -5.787e+01 | 0.0000 | 1503.0 | 5.82% | 1133.1 | 4.42% | motif file (matrix) |
pdf |
| 58 |  | MafA(bZIP)/Islet-MafA-ChIP-Seq(GSE30298)/Homer | 1e-25 | -5.782e+01 | 0.0000 | 2447.0 | 9.48% | 1970.5 | 7.69% | motif file (matrix) |
pdf |
| 59 |  | Rfx5(HTH)/GM12878-Rfx5-ChIP-Seq(GSE31477)/Homer | 1e-24 | -5.745e+01 | 0.0000 | 1059.0 | 4.10% | 754.2 | 2.94% | motif file (matrix) |
pdf |
| 60 |  | GATA:SCL/Ter119-SCL-ChIP-Seq(GSE18720)/Homer | 1e-24 | -5.556e+01 | 0.0000 | 414.0 | 1.60% | 238.7 | 0.93% | motif file (matrix) |
pdf |
| 61 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-22 | -5.197e+01 | 0.0000 | 4737.0 | 18.35% | 4113.5 | 16.06% | motif file (matrix) |
pdf |
| 62 |  | c-Jun-CRE(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-22 | -5.144e+01 | 0.0000 | 804.0 | 3.11% | 555.1 | 2.17% | motif file (matrix) |
pdf |
| 63 | 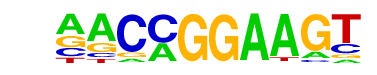 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-20 | -4.800e+01 | 0.0000 | 4179.0 | 16.19% | 3612.0 | 14.10% | motif file (matrix) |
pdf |
| 64 |  | EBF1(EBF)/Near-E2A-ChIP-Seq(GSE21512)/Homer | 1e-20 | -4.733e+01 | 0.0000 | 3518.0 | 13.63% | 3000.0 | 11.71% | motif file (matrix) |
pdf |
| 65 |  | AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-20 | -4.634e+01 | 0.0000 | 1693.0 | 6.56% | 1338.3 | 5.22% | motif file (matrix) |
pdf |
| 66 |  | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-19 | -4.477e+01 | 0.0000 | 3414.0 | 13.23% | 2916.8 | 11.38% | motif file (matrix) |
pdf |
| 67 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-19 | -4.443e+01 | 0.0000 | 2848.0 | 11.03% | 2394.9 | 9.35% | motif file (matrix) |
pdf |
| 68 |  | Rbpj1(?)/Panc1-Rbpj1-ChIP-Seq(GSE47459)/Homer | 1e-17 | -4.050e+01 | 0.0000 | 3906.0 | 15.13% | 3400.0 | 13.27% | motif file (matrix) |
pdf |
| 69 |  | NeuroD1(bHLH)/Islet-NeuroD1-ChIP-Seq(GSE30298)/Homer | 1e-16 | -3.839e+01 | 0.0000 | 2434.0 | 9.43% | 2043.4 | 7.98% | motif file (matrix) |
pdf |
| 70 |  | SPDEF(ETS)/VCaP-SPDEF-ChIP-Seq(SRA014231)/Homer | 1e-16 | -3.788e+01 | 0.0000 | 3011.0 | 11.67% | 2579.5 | 10.07% | motif file (matrix) |
pdf |
| 71 |  | Pax7-long(Paired/Homeobox)/Myoblast-Pax7-ChIP-Seq(GSE25064)/Homer | 1e-16 | -3.760e+01 | 0.0000 | 62.0 | 0.24% | 17.7 | 0.07% | motif file (matrix) |
pdf |
| 72 |  | Nur77(NR)/K562-NR4A1-ChIP-Seq(GSE31363)/Homer | 1e-15 | -3.666e+01 | 0.0000 | 498.0 | 1.93% | 335.6 | 1.31% | motif file (matrix) |
pdf |
| 73 |  | Ap4(HLH)/AML-Tfap4-ChIP-Seq(GSE45738)/Homer | 1e-14 | -3.390e+01 | 0.0000 | 3870.0 | 14.99% | 3408.7 | 13.30% | motif file (matrix) |
pdf |
| 74 |  | JunD(bZIP)/K562-JunD-ChIP-Seq/Homer | 1e-14 | -3.383e+01 | 0.0000 | 285.0 | 1.10% | 171.4 | 0.67% | motif file (matrix) |
pdf |
| 75 |  | Oct4:Sox17/F9-Sox17-ChIP-Seq(GSE44553)/Homer | 1e-14 | -3.304e+01 | 0.0000 | 340.0 | 1.32% | 215.1 | 0.84% | motif file (matrix) |
pdf |
| 76 |  | Atf1(bZIP)/K562-ATF1-ChIP-Seq(GSE31477)/Homer | 1e-14 | -3.266e+01 | 0.0000 | 1494.0 | 5.79% | 1213.8 | 4.74% | motif file (matrix) |
pdf |
| 77 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-14 | -3.265e+01 | 0.0000 | 1815.0 | 7.03% | 1504.1 | 5.87% | motif file (matrix) |
pdf |
| 78 |  | TR4(NR/DR1)/Hela-TR4-ChIP-Seq(GSE24685)/Homer | 1e-14 | -3.238e+01 | 0.0000 | 395.0 | 1.53% | 260.7 | 1.02% | motif file (matrix) |
pdf |
| 79 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-13 | -3.136e+01 | 0.0000 | 1714.0 | 6.64% | 1418.6 | 5.54% | motif file (matrix) |
pdf |
| 80 |  | Pdx1(Homeobox)/Islet-Pdx1-ChIP-Seq(SRA008281)/Homer | 1e-13 | -3.123e+01 | 0.0000 | 1823.0 | 7.06% | 1518.9 | 5.93% | motif file (matrix) |
pdf |
| 81 |  | Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-13 | -3.015e+01 | 0.0000 | 554.0 | 2.15% | 396.6 | 1.55% | motif file (matrix) |
pdf |
| 82 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-12 | -2.973e+01 | 0.0000 | 1803.0 | 6.99% | 1508.0 | 5.89% | motif file (matrix) |
pdf |
| 83 |  | GFY-Staf/Promoters/Homer | 1e-12 | -2.962e+01 | 0.0000 | 166.0 | 0.64% | 88.6 | 0.35% | motif file (matrix) |
pdf |
| 84 |  | EKLF(Zf)/Erythrocyte-Klf1-ChIP-Seq(GSE20478)/Homer | 1e-12 | -2.880e+01 | 0.0000 | 679.0 | 2.63% | 506.5 | 1.98% | motif file (matrix) |
pdf |
| 85 |  | Tbx20(T-box)/Heart-Tbx20-ChIP-Seq(GSE29636)/Homer | 1e-12 | -2.817e+01 | 0.0000 | 707.0 | 2.74% | 532.2 | 2.08% | motif file (matrix) |
pdf |
| 86 |  | Tbox:Smad/ESCd5-Smad2_3-ChIP-Seq(GSE29422)/Homer | 1e-12 | -2.764e+01 | 0.0000 | 590.0 | 2.29% | 433.7 | 1.69% | motif file (matrix) |
pdf |
| 87 |  | NFkB-p65(RHD)/GM12787-p65-ChIP-Seq(GSE19485)/Homer | 1e-11 | -2.690e+01 | 0.0000 | 1244.0 | 4.82% | 1012.5 | 3.95% | motif file (matrix) |
pdf |
| 88 |  | Olig2(bHLH)/Neuron-Olig2-ChIP-Seq(GSE30882)/Homer | 1e-11 | -2.561e+01 | 0.0000 | 4998.0 | 19.36% | 4545.7 | 17.74% | motif file (matrix) |
pdf |
| 89 |  | OCT4-SOX2-TCF-NANOG((POU/Homeobox/HMG)/mES-Oct4-ChIP-Seq(GSE11431)/Homer | 1e-10 | -2.445e+01 | 0.0000 | 314.0 | 1.22% | 210.6 | 0.82% | motif file (matrix) |
pdf |
| 90 |  | CRE(bZIP)/Promoter/Homer | 1e-10 | -2.422e+01 | 0.0000 | 588.0 | 2.28% | 441.0 | 1.72% | motif file (matrix) |
pdf |
| 91 |  | ETS(ETS)/Promoter/Homer | 1e-10 | -2.405e+01 | 0.0000 | 1114.0 | 4.32% | 907.3 | 3.54% | motif file (matrix) |
pdf |
| 92 |  | RUNX2(Runt)/PCa-RUNX2-ChIP-Seq(GSE33889)/Homer | 1e-10 | -2.396e+01 | 0.0000 | 1979.0 | 7.67% | 1700.4 | 6.64% | motif file (matrix) |
pdf |
| 93 |  | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-10 | -2.351e+01 | 0.0000 | 3491.0 | 13.53% | 3124.4 | 12.19% | motif file (matrix) |
pdf |
| 94 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-10 | -2.342e+01 | 0.0000 | 1725.0 | 6.68% | 1468.5 | 5.73% | motif file (matrix) |
pdf |
| 95 |  | EBF(EBF)/proBcell-EBF-ChIP-Seq(GSE21978)/Homer | 1e-10 | -2.309e+01 | 0.0000 | 750.0 | 2.91% | 586.7 | 2.29% | motif file (matrix) |
pdf |
| 96 |  | GATA-IR3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-9 | -2.272e+01 | 0.0000 | 306.0 | 1.19% | 207.7 | 0.81% | motif file (matrix) |
pdf |
| 97 | 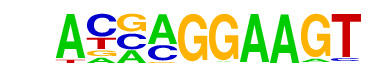 | ELF5(ETS)/T47D-ELF5-ChIP-Seq(GSE30407)/Homer | 1e-9 | -2.265e+01 | 0.0000 | 1990.0 | 7.71% | 1718.8 | 6.71% | motif file (matrix) |
pdf |
| 98 |  | FOXA1:AR/LNCAP-AR-ChIP-Seq(GSE27824)/Homer | 1e-9 | -2.231e+01 | 0.0000 | 139.0 | 0.54% | 77.7 | 0.30% | motif file (matrix) |
pdf |
| 99 |  | Bach2(bZIP)/OCILy7-Bach2-ChIP-Seq(GSE44420)/Homer | 1e-9 | -2.215e+01 | 0.0000 | 481.0 | 1.86% | 355.2 | 1.39% | motif file (matrix) |
pdf |
| 100 |  | EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer | 1e-9 | -2.213e+01 | 0.0000 | 1837.0 | 7.12% | 1579.5 | 6.16% | motif file (matrix) |
pdf |
| 101 |  | RUNX-AML(Runt)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer | 1e-9 | -2.166e+01 | 0.0000 | 1820.0 | 7.05% | 1566.7 | 6.12% | motif file (matrix) |
pdf |
| 102 |  | GATA-DR4(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-9 | -2.075e+01 | 0.0000 | 176.0 | 0.68% | 107.4 | 0.42% | motif file (matrix) |
pdf |
| 103 |  | Gfi1b(Zf)/HPC7-Gfi1b-ChIP-Seq(GSE22178)/Homer | 1e-8 | -2.049e+01 | 0.0000 | 1508.0 | 5.84% | 1285.0 | 5.02% | motif file (matrix) |
pdf |
| 104 |  | Erra(NR)/HepG2-Erra-ChIP-Seq(GSE31477)/Homer | 1e-8 | -2.006e+01 | 0.0000 | 5842.0 | 22.63% | 5412.2 | 21.12% | motif file (matrix) |
pdf |
| 105 |  | Six1(Homeobox)/Myoblast-Six1-ChIP-Chip(GSE20150)/Homer | 1e-8 | -1.947e+01 | 0.0000 | 540.0 | 2.09% | 414.3 | 1.62% | motif file (matrix) |
pdf |
| 106 |  | E-box(HLH)/Promoter/Homer | 1e-8 | -1.946e+01 | 0.0000 | 219.0 | 0.85% | 143.8 | 0.56% | motif file (matrix) |
pdf |
| 107 |  | HNF4a(NR/DR1)/HepG2-HNF4a-ChIP-Seq(GSE25021)/Homer | 1e-8 | -1.932e+01 | 0.0000 | 1286.0 | 4.98% | 1086.3 | 4.24% | motif file (matrix) |
pdf |
| 108 |  | Myf5(bHLH)/GM-Myf5-ChIP-Seq(GSE24852)/Homer | 1e-8 | -1.916e+01 | 0.0000 | 2249.0 | 8.71% | 1983.4 | 7.74% | motif file (matrix) |
pdf |
| 109 |  | Pax7(Paired/Homeobox)/Myoblast-Pax7-ChIP-Seq(GSE25064)/Homer | 1e-8 | -1.878e+01 | 0.0000 | 172.0 | 0.67% | 107.5 | 0.42% | motif file (matrix) |
pdf |
| 110 |  | Pbx3(Homeobox)/GM12878-PBX3-ChIP-Seq(GSE32465)/Homer | 1e-7 | -1.837e+01 | 0.0000 | 738.0 | 2.86% | 593.2 | 2.32% | motif file (matrix) |
pdf |
| 111 |  | Hoxb4(Homeobox)/ES-Hoxb4-ChIP-Seq(GSE34014)/Homer | 1e-7 | -1.802e+01 | 0.0000 | 416.0 | 1.61% | 311.9 | 1.22% | motif file (matrix) |
pdf |
| 112 |  | MyoG(HLH)/C2C12-MyoG-ChIP-Seq(GSE36024)/Homer | 1e-7 | -1.787e+01 | 0.0000 | 3413.0 | 13.22% | 3097.7 | 12.09% | motif file (matrix) |
pdf |
| 113 |  | GRHL2(CP2)/HBE-GRHL2-ChIP-Seq(GSE46194)/Homer | 1e-7 | -1.743e+01 | 0.0000 | 847.0 | 3.28% | 695.6 | 2.71% | motif file (matrix) |
pdf |
| 114 |  | RUNX1(Runt)/Jurkat-RUNX1-ChIP-Seq(GSE29180)/Homer | 1e-7 | -1.722e+01 | 0.0000 | 2524.0 | 9.78% | 2257.3 | 8.81% | motif file (matrix) |
pdf |
| 115 |  | PU.1(ETS)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-7 | -1.673e+01 | 0.0000 | 1343.0 | 5.20% | 1153.0 | 4.50% | motif file (matrix) |
pdf |
| 116 |  | Egr1(Zf)/K562-Egr1-ChIP-Seq(GSE32465)/Homer | 1e-7 | -1.669e+01 | 0.0000 | 2095.0 | 8.12% | 1856.1 | 7.24% | motif file (matrix) |
pdf |
| 117 |  | Nkx3.1(Homeobox)/LNCaP-Nkx3.1-ChIP-Seq(GSE28264)/Homer | 1e-7 | -1.642e+01 | 0.0000 | 5706.0 | 22.11% | 5321.9 | 20.77% | motif file (matrix) |
pdf |
| 118 |  | Tlx?(NR)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-6 | -1.587e+01 | 0.0000 | 1158.0 | 4.49% | 987.2 | 3.85% | motif file (matrix) |
pdf |
| 119 |  | Bcl6(Zf)/Liver-Bcl6-ChIP-Seq(GSE31578)/Homer | 1e-6 | -1.517e+01 | 0.0000 | 3306.0 | 12.81% | 3021.0 | 11.79% | motif file (matrix) |
pdf |
| 120 |  | GATA-IR4(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-6 | -1.483e+01 | 0.0000 | 177.0 | 0.69% | 118.4 | 0.46% | motif file (matrix) |
pdf |
| 121 |  | USF1(HLH)/GM12878-Usf1-ChIP-Seq(GSE32465)/Homer | 1e-6 | -1.468e+01 | 0.0000 | 989.0 | 3.83% | 838.6 | 3.27% | motif file (matrix) |
pdf |
| 122 |  | Usf2(HLH)/C2C12-Usf2-ChIP-Seq(GSE36030)/Homer | 1e-6 | -1.428e+01 | 0.0000 | 703.0 | 2.72% | 579.3 | 2.26% | motif file (matrix) |
pdf |
| 123 |  | RUNX(Runt)/HPC7-Runx1-ChIP-Seq(GSE22178)/Homer | 1e-5 | -1.362e+01 | 0.0000 | 1753.0 | 6.79% | 1557.1 | 6.08% | motif file (matrix) |
pdf |
| 124 |  | MafK(bZIP)/C2C12-MafK-ChIP-Seq(GSE36030)/Homer | 1e-5 | -1.356e+01 | 0.0000 | 612.0 | 2.37% | 500.6 | 1.95% | motif file (matrix) |
pdf |
| 125 |  | PBX1(Homeobox)/MCF7-PBX1-ChIP-Seq(GSE28007)/Homer | 1e-5 | -1.315e+01 | 0.0000 | 274.0 | 1.06% | 203.9 | 0.80% | motif file (matrix) |
pdf |
| 126 |  | MyoD(HLH)/Myotube-MyoD-ChIP-Seq(GSE21614)/Homer | 1e-5 | -1.277e+01 | 0.0000 | 2537.0 | 9.83% | 2308.8 | 9.01% | motif file (matrix) |
pdf |
| 127 |  | GFY(?)/Promoter/Homer | 1e-5 | -1.260e+01 | 0.0000 | 217.0 | 0.84% | 156.8 | 0.61% | motif file (matrix) |
pdf |
| 128 |  | Nkx2.5(Homeobox)/HL1-Nkx2.5.biotin-ChIP-Seq(GSE21529)/Homer | 1e-5 | -1.180e+01 | 0.0000 | 5617.0 | 21.76% | 5294.8 | 20.67% | motif file (matrix) |
pdf |
| 129 |  | PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-4 | -1.148e+01 | 0.0000 | 2988.0 | 11.58% | 2753.2 | 10.75% | motif file (matrix) |
pdf |
| 130 |  | NFkB-p65-Rel(RHD)/LPS-exp(GSE23622)/Homer | 1e-4 | -1.115e+01 | 0.0000 | 136.0 | 0.53% | 92.9 | 0.36% | motif file (matrix) |
pdf |
| 131 |  | Smad3(MAD)/NPC-Smad3-ChIP-Seq(GSE36673)/Homer | 1e-4 | -1.104e+01 | 0.0000 | 7908.0 | 30.64% | 7546.1 | 29.45% | motif file (matrix) |
pdf |
| 132 |  | Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer | 1e-4 | -1.063e+01 | 0.0000 | 889.0 | 3.44% | 769.2 | 3.00% | motif file (matrix) |
pdf |
| 133 |  | LXRE(NR/DR4)/BLRP(RAW)-LXRb-ChIP-Seq(GSE21512)/Homer | 1e-4 | -1.053e+01 | 0.0000 | 132.0 | 0.51% | 90.8 | 0.35% | motif file (matrix) |
pdf |
| 134 |  | AMYB(HTH)/Testes-AMYB-ChIP-Seq(GSE44588)/Homer | 1e-4 | -1.035e+01 | 0.0001 | 3910.0 | 15.15% | 3656.5 | 14.27% | motif file (matrix) |
pdf |
| 135 |  | Atoh1(bHLH)/Cerebellum-Atoh1-ChIP-Seq(GSE22111)/Homer | 1e-4 | -1.012e+01 | 0.0001 | 3138.0 | 12.16% | 2913.7 | 11.37% | motif file (matrix) |
pdf |
| 136 |  | PPARE(NR/DR1)/3T3L1-Pparg-ChIP-Seq(GSE13511)/Homer | 1e-4 | -9.539e+00 | 0.0001 | 2546.0 | 9.86% | 2350.5 | 9.17% | motif file (matrix) |
pdf |
| 137 |  | PAX3:FKHR-fusion(Paired/Homeobox)/Rh4-PAX3:FKHR-ChIP-Seq(GSE19063)/Homer | 1e-3 | -9.084e+00 | 0.0002 | 364.0 | 1.41% | 296.7 | 1.16% | motif file (matrix) |
pdf |
| 138 |  | RXR(NR/DR1)/3T3L1-RXR-ChIP-Seq(GSE13511)/Homer | 1e-3 | -9.038e+00 | 0.0002 | 3091.0 | 11.98% | 2881.5 | 11.25% | motif file (matrix) |
pdf |
| 139 |  | Egr2/Thymocytes-Egr2-ChIP-Seq(GSE34254)/Homer | 1e-3 | -8.497e+00 | 0.0004 | 503.0 | 1.95% | 425.8 | 1.66% | motif file (matrix) |
pdf |
| 140 |  | Bach1(bZIP)/K562-Bach1-ChIP-Seq(GSE31477)/Homer | 1e-3 | -8.423e+00 | 0.0004 | 112.0 | 0.43% | 78.4 | 0.31% | motif file (matrix) |
pdf |
| 141 |  | BMYB(HTH)/Hela-BMYB-ChIPSeq(GSE27030)/Homer | 1e-3 | -8.418e+00 | 0.0004 | 3766.0 | 14.59% | 3543.1 | 13.83% | motif file (matrix) |
pdf |
| 142 |  | PAX5-shortForm(Paired/Homeobox)/GM12878-PAX5-ChIP-Seq(GSE32465)/Homer | 1e-3 | -8.364e+00 | 0.0004 | 251.0 | 0.97% | 198.2 | 0.77% | motif file (matrix) |
pdf |
| 143 |  | Nrf2(bZIP)/Lymphoblast-Nrf2-ChIP-Seq(GSE37589)/Homer | 1e-3 | -8.137e+00 | 0.0005 | 104.0 | 0.40% | 72.6 | 0.28% | motif file (matrix) |
pdf |
| 144 |  | E2F6(E2F)/Hela-E2F6-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.624e+00 | 0.0008 | 1561.0 | 6.05% | 1427.2 | 5.57% | motif file (matrix) |
pdf |
| 145 |  | PRDM1/BMI1(Zf)/Hela-PRDM1-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.618e+00 | 0.0008 | 1113.0 | 4.31% | 1001.1 | 3.91% | motif file (matrix) |
pdf |
| 146 |  | IRF4(IRF)/GM12878-IRF4-ChIP-Seq(GSE32465)/Homer | 1e-3 | -7.594e+00 | 0.0008 | 869.0 | 3.37% | 771.9 | 3.01% | motif file (matrix) |
pdf |
| 147 |  | Nkx2.1(Homeobox)/LungAC-Nkx2.1-ChIP-Seq(GSE43252)/Homer | 1e-3 | -7.273e+00 | 0.0012 | 7283.0 | 28.22% | 7001.6 | 27.33% | motif file (matrix) |
pdf |
| 148 |  | NRF1(NRF)/MCF7-NRF1-ChIP-Seq(Unpublished)/Homer | 1e-2 | -6.501e+00 | 0.0025 | 453.0 | 1.76% | 390.5 | 1.52% | motif file (matrix) |
pdf |
| 149 |  | Stat3(Stat)/mES-Stat3-ChIP-Seq(GSE11431)/Homer | 1e-2 | -6.470e+00 | 0.0026 | 1246.0 | 4.83% | 1138.1 | 4.44% | motif file (matrix) |
pdf |
| 150 |  | Pax8(Paired/Homeobox)/Thyroid-Pax8-ChIP-Seq(GSE26938)/Homer | 1e-2 | -6.439e+00 | 0.0026 | 765.0 | 2.96% | 682.7 | 2.66% | motif file (matrix) |
pdf |
| 151 |  | NRF1/Promoter/Homer | 1e-2 | -6.430e+00 | 0.0026 | 500.0 | 1.94% | 434.7 | 1.70% | motif file (matrix) |
pdf |
| 152 |  | Esrrb(NR)/mES-Esrrb-ChIP-Seq(GSE11431)/Homer | 1e-2 | -6.225e+00 | 0.0032 | 1513.0 | 5.86% | 1396.0 | 5.45% | motif file (matrix) |
pdf |
| 153 |  | Tcfcp2l1(CP2)/mES-Tcfcp2l1-ChIP-Seq(GSE11431)/Homer | 1e-2 | -5.982e+00 | 0.0041 | 427.0 | 1.65% | 369.5 | 1.44% | motif file (matrix) |
pdf |
| 154 |  | TATA-Box(TBP)/Promoter/Homer | 1e-2 | -5.732e+00 | 0.0052 | 2627.0 | 10.18% | 2478.6 | 9.67% | motif file (matrix) |
pdf |
| 155 |  | MYB(HTH)/ERMYB-Myb-ChIPSeq(GSE22095)/Homer | 1e-2 | -5.622e+00 | 0.0057 | 4343.0 | 16.83% | 4152.2 | 16.21% | motif file (matrix) |
pdf |
| 156 |  | Znf263(Zf)/K562-Znf263-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.501e+00 | 0.0064 | 5644.0 | 21.87% | 5430.0 | 21.19% | motif file (matrix) |
pdf |
| 157 |  | Stat3+il21(Stat)/CD4-Stat3-ChIP-Seq(GSE19198)/Homer | 1e-2 | -5.488e+00 | 0.0065 | 1719.0 | 6.66% | 1603.7 | 6.26% | motif file (matrix) |
pdf |
| 158 |  | NF-E2(bZIP)/K562-NFE2-ChIP-Seq(GSE31477)/Homer | 1e-2 | -4.996e+00 | 0.0105 | 120.0 | 0.46% | 94.3 | 0.37% | motif file (matrix) |
pdf |
| 159 |  | GATA-DR8(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-2 | -4.760e+00 | 0.0132 | 127.0 | 0.49% | 101.4 | 0.40% | motif file (matrix) |
pdf |
| 160 |  | ZNF143|STAF(Zf)/CUTLL-ZNF143-ChIP-Seq(GSE29600)/Homer | 1e-2 | -4.732e+00 | 0.0135 | 953.0 | 3.69% | 876.1 | 3.42% | motif file (matrix) |
pdf |































































































































































































































































































































